HDX workbench: software for the analysis of H/D exchange MS data - PubMed (original) (raw)
HDX workbench: software for the analysis of H/D exchange MS data
Bruce D Pascal et al. J Am Soc Mass Spectrom. 2012 Sep.
Abstract
Hydrogen/deuterium exchange mass spectrometry (HDX-MS) is an established method for the interrogation of protein conformation and dynamics. While the data analysis challenge of HDX-MS has been addressed by a number of software packages, new computational tools are needed to keep pace with the improved methods and throughput of this technique. To address these needs, we report an integrated desktop program titled HDX Workbench, which facilitates automation, management, visualization, and statistical cross-comparison of large HDX data sets. Using the software, validated data analysis can be achieved at the rate of generation. The application is available at the project home page http://hdx.florida.scripps.edu .
Figures
Figure 1
The standardized workflow for HDX analysis outlines the common steps for data analysis of an HDX experiment. This workflow has been established over several years and has driven the design of the software
Figure 2
The protein editor allows for the creation and editing of all aspects of a protein used in HDX analysis. Sequence, peptide set, and secondary structure features can be managed using this interface. The protein configuration can be saved for reuse or shared with other users
Figure 3
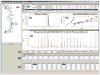
The Perturbation View provides a central feature set for integrating several graphic tools for editing and review of HDX data. (a) Tree pane is the primary launch point of the software which allows users to manage project and experiment data. A wide range of functions are available contextually from each node such as launching a detect job, editing experiment information, editing protein information, and loading result data. Only projects associated with the specific user are shown. (b) Peptide summary table provides experiment level information for each peptide. Peptide selection will launch results in the subsequent tools. (c) The extracted ion chromatogram (XIC) from selected replicates are color coded and displayed in a single view, facilitating validation of the peptide assignments and providing feedback regarding chromatographic consistency over several runs. (d) Deuterium uptake plots display the perturbation data for all time point replicates in one view, and are updated automatically. The results of _t_-tests between samples are shown above each time point to illustrate statistical significance. Also shown above are the intrinsic exchange rate plot (black) for the selected peptide calculated based on the work from Walter Englander. (e) Spectra pane displays the co-added raw spectral data for one or many replicates. Sub-range bars are colored grey. Shown above are the spectra for Apo replicate 1 at 0 and 3600 s rendered concurrently. (f) Information toolbar allows recalculation of input values for centroiding approach and displays additional information. Tables (g) and (h) allow users to load one to many replicates in the spectral and XIC panes for review. The replicate table (h) displays %D results for each time point replicate determined by both centroid and theoretical methods. Right clicking a cell will discard the replicate from the data set, automatically updating all views. (c), (d) (e) Support zoom in/out functionality
Figure 4
Sequence coverage view. Panel (a) shows heat map coverage plots for a single sample. The level of deuterium incorporation %D for each time point is rendered in colored horizontal layers for each peptide, allowing for visualization of the rate of exchange in a single block. Panel (b) displays colored differential HDX data rendered onto the protein sequence. The difference level of deuterium incorporation measured as a percentage (delta %D), and it associated standard deviation are displayed on each peptide to facilitate the validation process. The bar at the beginning of each peptide represents the first two residues in which the exchange rate occurs too quickly to be measured. The secondary structure elements are rendered automatically from the features defined in the protein editor and provide additional context to the peptide coverage
Figure 5
Experiment comparison. This tool allows cross comparison of result data from multiple experiments. Shown above is perturbation data for Vitamin D receptor in the presence of 31 ligands displayed in a single view. Cells are colored according to a heat map legend, facilitating visual identification of regions of stability or exposure. Column selection, sorting, and formatting can be adjusted or ordered according to similarity. Peptides can be removed if not present in all experiments. Results can be exported from this view in text or graphical format. Right clicking a peptide will launch statistical analysis features in which the user may select a representative time point, and the results from all experiments for the selected peptide/ time point combination will be compared for statistical significance using one-way ANOVA and Tukey’s multiple comparison
Similar articles
- HDX-analyzer: a novel package for statistical analysis of protein structure dynamics.
Liu S, Liu L, Uzuner U, Zhou X, Gu M, Shi W, Zhang Y, Dai SY, Yuan JS. Liu S, et al. BMC Bioinformatics. 2011 Feb 15;12 Suppl 1(Suppl 1):S43. doi: 10.1186/1471-2105-12-S1-S43. BMC Bioinformatics. 2011. PMID: 21342575 Free PMC article. - The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data.
Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. Pascal BD, et al. BMC Bioinformatics. 2007 May 16;8:156. doi: 10.1186/1471-2105-8-156. BMC Bioinformatics. 2007. PMID: 17506883 Free PMC article. - HD desktop: an integrated platform for the analysis and visualization of H/D exchange data.
Pascal BD, Chalmers MJ, Busby SA, Griffin PR. Pascal BD, et al. J Am Soc Mass Spectrom. 2009 Apr;20(4):601-10. doi: 10.1016/j.jasms.2008.11.019. Epub 2008 Dec 6. J Am Soc Mass Spectrom. 2009. PMID: 19135386 Free PMC article. - Computational Tools for Hydrogen-Deuterium Exchange Mass Spectrometry Data Analysis.
Stofella M, Grimaldi A, Smit JH, Claesen J, Paci E, Sobott F. Stofella M, et al. Chem Rev. 2024 Nov 13;124(21):12242-12263. doi: 10.1021/acs.chemrev.4c00438. Epub 2024 Oct 31. Chem Rev. 2024. PMID: 39481095 Free PMC article. Review. - Protein dynamics and conformational changes explored by hydrogen/deuterium exchange mass spectrometry.
Zheng J, Strutzenberg T, Pascal BD, Griffin PR. Zheng J, et al. Curr Opin Struct Biol. 2019 Oct;58:305-313. doi: 10.1016/j.sbi.2019.06.007. Epub 2019 Jul 25. Curr Opin Struct Biol. 2019. PMID: 31351767 Review.
Cited by
- Molecular insights into frataxin-mediated iron supply for heme biosynthesis in Bacillus subtilis.
Mielcarek A, Blauenburg B, Miethke M, Marahiel MA. Mielcarek A, et al. PLoS One. 2015 Mar 31;10(3):e0122538. doi: 10.1371/journal.pone.0122538. eCollection 2015. PLoS One. 2015. PMID: 25826316 Free PMC article. - Structure of phosphorylated-like RssB, the adaptor delivering σs to the ClpXP proteolytic machinery, reveals an interface switch for activation.
Brugger C, Schwartz J, Novick S, Tong S, Hoskins JR, Majdalani N, Kim R, Filipovski M, Wickner S, Gottesman S, Griffin PR, Deaconescu AM. Brugger C, et al. J Biol Chem. 2023 Dec;299(12):105440. doi: 10.1016/j.jbc.2023.105440. Epub 2023 Nov 9. J Biol Chem. 2023. PMID: 37949227 Free PMC article. - DECA, A Comprehensive, Automatic Post-processing Program for HDX-MS Data.
Lumpkin RJ, Komives EA. Lumpkin RJ, et al. Mol Cell Proteomics. 2019 Dec;18(12):2516-2523. doi: 10.1074/mcp.TIR119.001731. Epub 2019 Oct 8. Mol Cell Proteomics. 2019. PMID: 31594786 Free PMC article. - Structural Basis of Altered Potency and Efficacy Displayed by a Major in Vivo Metabolite of the Antidiabetic PPARγ Drug Pioglitazone.
Mosure SA, Shang J, Eberhardt J, Brust R, Zheng J, Griffin PR, Forli S, Kojetin DJ. Mosure SA, et al. J Med Chem. 2019 Feb 28;62(4):2008-2023. doi: 10.1021/acs.jmedchem.8b01573. Epub 2019 Feb 7. J Med Chem. 2019. PMID: 30676741 Free PMC article. - Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Zhao LH, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Hou L, de Waal PW, Li S, Jiang Y, Scaffidi A, Flematti GR, Smith SM, Lam VQ, Griffin PR, Wang Y, Li J, Melcher K, Xu HE. Zhao LH, et al. Cell Res. 2015 Nov;25(11):1219-36. doi: 10.1038/cr.2015.122. Epub 2015 Oct 16. Cell Res. 2015. PMID: 26470846 Free PMC article.
References
- Abzalimov RR, Kaltashov IA. Extraction of Local Hydrogen Exchange Data from HDX CAD MS Measurements by Deconvolution of Isotopic Distributions of Fragment Ions. J Am Soc Mass Spectrom. 2006;17 (11):1543–1551. - PubMed
- Chik JK, Vande Graaf JL, Schriemer DC. Quantitating the statistical distribution of deuterium incorporation to extend the utility of H/D exchange MS data. Anal Chem. 2006;78(1):207–214. - PubMed
- Lou XH, Kirchner M, Renard BY, Kothe U, Boppel S, Graf C, Lee CT, Steen JAJ, Steen H, Mayer MP, Hamprecht FA. Deuteration Distribution Estimation with Improved Sequence Coverage for HX/MS Experiments. Bioinformatics. 2010;26:1535–1541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources