Structure, function and diversity of the healthy human microbiome - PubMed (original) (raw)
Structure, function and diversity of the healthy human microbiome
Human Microbiome Project Consortium. Nature. 2012.
Abstract
Studies of the human microbiome have revealed that even healthy individuals differ remarkably in the microbes that occupy habitats such as the gut, skin and vagina. Much of this diversity remains unexplained, although diet, environment, host genetics and early microbial exposure have all been implicated. Accordingly, to characterize the ecology of human-associated microbial communities, the Human Microbiome Project has analysed the largest cohort and set of distinct, clinically relevant body habitats so far. We found the diversity and abundance of each habitat's signature microbes to vary widely even among healthy subjects, with strong niche specialization both within and among individuals. The project encountered an estimated 81-99% of the genera, enzyme families and community configurations occupied by the healthy Western microbiome. Metagenomic carriage of metabolic pathways was stable among individuals despite variation in community structure, and ethnic/racial background proved to be one of the strongest associations of both pathways and microbes with clinical metadata. These results thus delineate the range of structural and functional configurations normal in the microbial communities of a healthy population, enabling future characterization of the epidemiology, ecology and translational applications of the human microbiome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. Diversity of the human microbiome is concordant among measures, unique to each individual, and strongly determined by microbial habitat
A) Alpha diversity within subjects by body habitat, as measured using the relative inverse Simpson index of 16S rRNA gene OTUs (red), genus-level phylotypes (blue), shotgun metagenomic reads matched to reference genomes (green), functional modules (yellow), and enzyme families (white). The mouth generally shows high within-subject diversity and the vagina low diversity, with other habitats intermediate; variation among individuals often exceeds variation among body habitats. B) Bray-Curtis beta diversity among subjects by body habitat, colors as for A. Skin differs most between subjects, with oral habitats and vaginal genera more stable. Although alpha- and beta-diversity are not directly comparable, changes in structure among communities (A) occupy a wider dynamic range than do changes within communities among individuals (B). C) Principal coordinates plot showing variation among samples demonstrates that primary clustering is by body area, with the oral, gastrointestinal, skin, and urogenital habitats separate; the nares bridge oral and skin habitats. D) Repeated samples from the same subject (red) are more similar than microbiomes from different subjects (yellow). Technical replicates (green) are in turn more similar; these patterns are consistent for all body habitats and for both phylogenetic and metabolic community composition. See previously described sample counts for all comparisons.
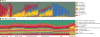
Figure 2. Carriage of microbial taxa varies while metabolic pathways remain stable within a healthy population
Vertical bars represent microbiome samples by body habitat in the seven locations with both shotgun and 16S data; bars indicate relative abundances colored by A) microbial phyla from binned OTUs and B) metabolic modules. Legend indicates most abundant phyla/pathways by average within one or more body habitats; RC = retroauricular crease. A plurality of most communities’ memberships consists of a single dominant phylum (and often genus; see Supp. Fig. 2), but this is universal neither to all body habitats nor to all individuals. Conversely, most metabolic pathways are evenly distributed and prevalent across both individuals and body habitats.
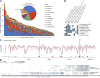
Figure 3. Abundant taxa in the human microbiome, which has been metagenomically and taxonomically well-defined in the HMP population
A–C) Prevalence (intensity, color denoting phylum/class) and abundance when present (size) of clades in the healthy microbiome. The most abundant A) metagenomically-identified species, B) 16S-identified genera, and C) PATRIC “pathogens” (metagenomic). The population size and sequencing depths of the HMP have well-defined the microbiome at all assayed body sites, as assessed by saturation of added community D) metabolic configurations (rarefaction of minimum Bray-Curtis β-diversity of metagenomic EC abundances to nearest neighbor, inter-quartile range over 100 samples) and E) phylogenetic configurations (min. 16S OTU weighted UniFrac distance to nearest neighbor).
Figure 4. Microbial carriage varies between subjects down to the species and strain level
Metagenomic reads from 127 tongue samples spanning 90 subjects were processed with MetaPhlAn to determine relative abundances for each species. A) Relative abundances of 11 distinct Streptococcus spp. In addition to variation in broader clades (see Fig. 2), individual species within a single habitat demonstrate a wide range of compositional variation. Inset illustrates average tongue sample composition. B) Metabolic modules present/absent (grey/white) in KEGG reference genomes of tongue streptococci denote selected areas of strain-specific functional differentiation. C) Comparative genomic coverage for the single Streptococcus mitis B6 strain. Grey dots are median Reads Per Kilobase per Million reads (RPKM) for 1kb windows, gray bars are the 25th to 75th percentiles across all samples, red line the lowess smoothed average. Red bars at the bottom highlight predicted genomic islands. Large, discrete, and highly variable islands are commonly under-represented. D) Two islands highlighted, V = V-type H+ ATPase subunits I,K,E,C,F,A & B, and CH = Choline binding proteins cbp6 and cbp12, indicating functional cohesion of strain-specific gene loss within individual human hosts.
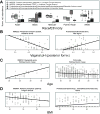
Figure 5. Microbial community membership and function correlates with host phenotype and sample metadata
The pathway and clade abundances most significantly associated (all FDR q<0.2) using a multivariate linear model with A) subject race or ethnicity, B) vaginal posterior fornix pH, C) subject age, and D) BMI. Samples’ scatter plots are shown with lines indicating best simple linear fit. Race/ethnicity and vaginal pH are particularly strong associations; age and BMI are more representative of typically modest phenotypic associations (Sup. Table 3), suggesting that variation in the healthy microbiota may correspond to other host or environmental factors.
Comment in
- Microbiology: Learning about who we are.
Relman DA. Relman DA. Nature. 2012 Jun 13;486(7402):194-5. doi: 10.1038/486194a. Nature. 2012. PMID: 22699602 No abstract available. - The human microbiome: there is much left to do.
Ley R. Ley R. Nature. 2022 Jun;606(7914):435. doi: 10.1038/d41586-022-01610-5. Nature. 2022. PMID: 35705822 No abstract available.
Similar articles
- A framework for human microbiome research.
Human Microbiome Project Consortium. Human Microbiome Project Consortium. Nature. 2012 Jun 13;486(7402):215-21. doi: 10.1038/nature11209. Nature. 2012. PMID: 22699610 Free PMC article. - Covariation of the Fecal Microbiome with Diet in Nonpasserine Birds.
Xiao K, Fan Y, Zhang Z, Shen X, Li X, Liang X, Bi R, Wu Y, Zhai J, Dai J, Irwin DM, Chen W, Shen Y. Xiao K, et al. mSphere. 2021 May 12;6(3):e00308-21. doi: 10.1128/mSphere.00308-21. mSphere. 2021. PMID: 33980682 Free PMC article. - Microbial diversity and interactions in subgingival biofilm communities.
Diaz PI. Diaz PI. Front Oral Biol. 2012;15:17-40. doi: 10.1159/000329669. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142955 Review. - Enterotypes in the landscape of gut microbial community composition.
Costea PI, Hildebrand F, Arumugam M, Bäckhed F, Blaser MJ, Bushman FD, de Vos WM, Ehrlich SD, Fraser CM, Hattori M, Huttenhower C, Jeffery IB, Knights D, Lewis JD, Ley RE, Ochman H, O'Toole PW, Quince C, Relman DA, Shanahan F, Sunagawa S, Wang J, Weinstock GM, Wu GD, Zeller G, Zhao L, Raes J, Knight R, Bork P. Costea PI, et al. Nat Microbiol. 2018 Jan;3(1):8-16. doi: 10.1038/s41564-017-0072-8. Epub 2017 Dec 18. Nat Microbiol. 2018. PMID: 29255284 Free PMC article. Review.
Cited by
- Serum Microcystin-LR Levels Linked with Risk of Inflammatory Bowel Disease: A Matched Case-Control Study in China.
Chen X, Liu W, Liao Y, Xiang X, Yang Y, Tian L, Gao X, Cao Q, Guo H, Chao K, Song F, Liu R, Li Y, Ye S, Wang J, Yang F, Wang X. Chen X, et al. Environ Health (Wash). 2024 May 3;2(7):453-464. doi: 10.1021/envhealth.3c00212. eCollection 2024 Jul 19. Environ Health (Wash). 2024. PMID: 39473886 Free PMC article. - Immune mechanisms and the impact of the disrupted lung microbiome in chronic bacterial lung infection and bronchiectasis.
Boyton RJ, Reynolds CJ, Quigley KJ, Altmann DM. Boyton RJ, et al. Clin Exp Immunol. 2013 Feb;171(2):117-23. doi: 10.1111/cei.12003. Clin Exp Immunol. 2013. PMID: 23286938 Free PMC article. Review. - Categorization of the gut microbiota: enterotypes or gradients?
Jeffery IB, Claesson MJ, O'Toole PW, Shanahan F. Jeffery IB, et al. Nat Rev Microbiol. 2012 Sep;10(9):591-2. doi: 10.1038/nrmicro2859. Nat Rev Microbiol. 2012. PMID: 23066529 No abstract available. - Microbiota and Tuberculosis: A Potential Role of Probiotics, and Postbiotics.
Liu Y, Wang J, Wu C. Liu Y, et al. Front Nutr. 2021 May 7;8:626254. doi: 10.3389/fnut.2021.626254. eCollection 2021. Front Nutr. 2021. PMID: 34026804 Free PMC article. Review. - Natural selection in a bangladeshi population from the cholera-endemic ganges river delta.
Karlsson EK, Harris JB, Tabrizi S, Rahman A, Shlyakhter I, Patterson N, O'Dushlaine C, Schaffner SF, Gupta S, Chowdhury F, Sheikh A, Shin OS, Ellis C, Becker CE, Stuart LM, Calderwood SB, Ryan ET, Qadri F, Sabeti PC, Larocque RC. Karlsson EK, et al. Sci Transl Med. 2013 Jul 3;5(192):192ra86. doi: 10.1126/scitranslmed.3006338. Sci Transl Med. 2013. PMID: 23825302 Free PMC article.
References
- The Human Microbiome Project Consortium. A framework for human microbiome research. (in review)
- Aagaard K, et al. A Comprehensive Strategy for Sampling the Human Microbiome. (in review)
Publication types
MeSH terms
Substances
Grants and funding
- R01HG004856/HG/NHGRI NIH HHS/United States
- R01HG004908/HG/NHGRI NIH HHS/United States
- P30DE020751/DE/NIDCR NIH HHS/United States
- R21HG005811/HG/NHGRI NIH HHS/United States
- R01HG005172/HG/NHGRI NIH HHS/United States
- UH3DK083993/DK/NIDDK NIH HHS/United States
- R01 HG005969/HG/NHGRI NIH HHS/United States
- R01HG004900/HG/NHGRI NIH HHS/United States
- UH2 DK083990/DK/NIDDK NIH HHS/United States
- N01HG62088/HG/NHGRI NIH HHS/United States
- R01 HG004857/HG/NHGRI NIH HHS/United States
- U54 HG004973/HG/NHGRI NIH HHS/United States
- U01DE016937/DE/NIDCR NIH HHS/United States
- UH2 AR057504/AR/NIAMS NIH HHS/United States
- U54HG004969/HG/NHGRI NIH HHS/United States
- T32 AI007528/AI/NIAID NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- R01HG004877/HG/NHGRI NIH HHS/United States
- R01 HG004900/HG/NHGRI NIH HHS/United States
- DP2OD001500/OD/NIH HHS/United States
- R01HG004885/HG/NHGRI NIH HHS/United States
- R01HG004872/HG/NHGRI NIH HHS/United States
- R01 HG004856/HG/NHGRI NIH HHS/United States
- R01 HG005171/HG/NHGRI NIH HHS/United States
- UH3AI083263/AI/NIAID NIH HHS/United States
- UH2 AR057506/AR/NIAMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- UH2AR057504/AR/NIAMS NIH HHS/United States
- U01HG004866/HG/NHGRI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- U54 AI084844/AI/NIAID NIH HHS/United States
- UH3AR057504/AR/NIAMS NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- UH2AI083263/AI/NIAID NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- U54AI084844/AI/NIAID NIH HHS/United States
- R01 HG005975/HG/NHGRI NIH HHS/United States
- R37 DE016937/DE/NIDCR NIH HHS/United States
- UH3 DK083993/DK/NIDDK NIH HHS/United States
- R01DE021574/DE/NIDCR NIH HHS/United States
- R01 HG004885/HG/NHGRI NIH HHS/United States
- U54 HG004968/HG/NHGRI NIH HHS/United States
- U54HG004973/HG/NHGRI NIH HHS/United States
- R01HG004857/HG/NHGRI NIH HHS/United States
- UH2 AI083263/AI/NIAID NIH HHS/United States
- R01HG005171/HG/NHGRI NIH HHS/United States
- T32AI007528/AI/NIAID NIH HHS/United States
- R01HG004906/HG/NHGRI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- RC1 DE020298/DE/NIDCR NIH HHS/United States
- DP2 OD001500/OD/NIH HHS/United States
- R01 DE021574/DE/NIDCR NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
- R01 HG004908/HG/NHGRI NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- R21 CA139193/CA/NCI NIH HHS/United States
- R01HG005975/HG/NHGRI NIH HHS/United States
- U01 DE016937/DE/NIDCR NIH HHS/United States
- R21CA139193/CA/NCI NIH HHS/United States
- R21 HG005811/HG/NHGRI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R01 HG004853/HG/NHGRI NIH HHS/United States
- N01AI30071/AI/NIAID NIH HHS/United States
- U54 HG004969/HG/NHGRI NIH HHS/United States
- R01HG004853/HG/NHGRI NIH HHS/United States
- UH3 AI083263/AI/NIAID NIH HHS/United States
- UH2DK083990/DK/NIDDK NIH HHS/United States
- U54HG004968/HG/NHGRI NIH HHS/United States
- RC1DE0202098/DE/NIDCR NIH HHS/United States
- R01 HG004906/HG/NHGRI NIH HHS/United States
- R01HG005969/HG/NHGRI NIH HHS/United States
- R01 HG004877/HG/NHGRI NIH HHS/United States
- P30 DE020751/DE/NIDCR NIH HHS/United States
- R01 HG005172/HG/NHGRI NIH HHS/United States
- UH2AR057506/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases