The VHL/HIF axis in clear cell renal carcinoma - PubMed (original) (raw)
Review
The VHL/HIF axis in clear cell renal carcinoma
Chuan Shen et al. Semin Cancer Biol. 2013 Feb.
Abstract
Inactivation of the VHL tumor suppressor protein (pVHL) is a common event in clear cell renal carcinoma, which is the most common form of kidney cancer. pVHL performs many functions, including serving as the substrate recognition module of an ubiquitin ligase complex that targets the alpha subunits of the heterodimeric HIF transcription factor for proteasomal degradation. Deregulation of HIF2α appears to be a driving force in pVHL-defective clear cell renal carcinomas. In contrast, genetic and functional studies suggest that HIF1α serves as a tumor suppressor and is a likely target of the 14q deletions that are characteristic of this tumor type. Drugs that inhibit HIF2α, or its downstream targets such as VEGF, are in various stages of clinical testing. Indeed, clear cell renal carcinomas are exquisitely sensitive to VEGF deprivation and four VEGF inhibitors have now been approved for the treatment of this disease.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
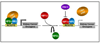
Fig 1. Role of pVHL, HIF1α, and HIF2α in Clear Cell Carcinoma
Loss of pVHL leads to the accumulation of both HIF1α and HIF2α, which have opposing effects on clear cell carcinogenesis. The leads to selection pressure to downregulate HIF1α.
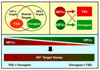
Fig 2. Quantitative differences between HIF1α and HIF2α related to activation of HIF target genes
HIF1α is more sensitive than HIF2α to inhibition by FIH1. Competitive displacement of HIF2α by HIF1α in a well-oxygenated pVHL-defective cell might therefore decrease the transcription of certain genes that are responsive to HIFα CTAD function.
Fig 3. Qualititative differences between HIF1α and HIF2α related to activation of HIF target genes
The genesets regulated by HIF1α and HIF2α overlap but are not entirely congruent. The former might be biased toward renal carcinoma suppressors and the latter to renal carcinoma oncoproteins.
Fig 4. Qualititative differences between HIF1α and HIF2α with respect to non-canonical HIF functions
HIF1α and HIF2α have opposing actions in some systems with respect to collateral signaling pathways involving cancer-relevant proteins such as c-Myc and p53.
Similar articles
- The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma.
Kaelin WG Jr. Kaelin WG Jr. Clin Cancer Res. 2007 Jan 15;13(2 Pt 2):680s-684s. doi: 10.1158/1078-0432.CCR-06-1865. Clin Cancer Res. 2007. PMID: 17255293 Review. - Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease.
Zatyka M, da Silva NF, Clifford SC, Morris MR, Wiesener MS, Eckardt KU, Houlston RS, Richards FM, Latif F, Maher ER. Zatyka M, et al. Cancer Res. 2002 Jul 1;62(13):3803-11. Cancer Res. 2002. PMID: 12097293 - The von Hippel-Lindau tumor suppressor gene and kidney cancer.
Kaelin WG Jr. Kaelin WG Jr. Clin Cancer Res. 2004 Sep 15;10(18 Pt 2):6290S-5S. doi: 10.1158/1078-0432.CCR-sup-040025. Clin Cancer Res. 2004. PMID: 15448019 Review. - von-Hippel Lindau and Hypoxia-Inducible Factor at the Center of Renal Cell Carcinoma Biology.
Shirole NH, Kaelin WG Jr. Shirole NH, et al. Hematol Oncol Clin North Am. 2023 Oct;37(5):809-825. doi: 10.1016/j.hoc.2023.04.011. Epub 2023 Jun 1. Hematol Oncol Clin North Am. 2023. PMID: 37270382 Free PMC article. Review. - Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1alpha in clear cell renal carcinomas.
Wiesener MS, Münchenhagen PM, Berger I, Morgan NV, Roigas J, Schwiertz A, Jürgensen JS, Gruber G, Maxwell PH, Löning SA, Frei U, Maher ER, Gröne HJ, Eckardt KU. Wiesener MS, et al. Cancer Res. 2001 Jul 1;61(13):5215-22. Cancer Res. 2001. PMID: 11431362
Cited by
- Candidate biomarkers for treatment benefit from sunitinib in patients with advanced renal cell carcinoma using mass spectrometry-based (phospho)proteomics.
van der Wijngaart H, Beekhof R, Knol JC, Henneman AA, de Goeij-de Haas R, Piersma SR, Pham TV, Jimenez CR, Verheul HMW, Labots M. van der Wijngaart H, et al. Clin Proteomics. 2023 Nov 8;20(1):49. doi: 10.1186/s12014-023-09437-6. Clin Proteomics. 2023. PMID: 37940875 Free PMC article. - Immune-restoring CAR-T cells display antitumor activity and reverse immunosuppressive TME in a humanized ccRCC mouse model.
Wang Y, Cho JW, Kastrunes G, Buck A, Razimbaud C, Culhane AC, Sun J, Braun DA, Choueiri TK, Wu CJ, Jones K, Nguyen QD, Zhu Z, Wei K, Zhu Q, Signoretti S, Freeman GJ, Hemberg M, Marasco WA. Wang Y, et al. iScience. 2024 Jan 15;27(2):108879. doi: 10.1016/j.isci.2024.108879. eCollection 2024 Feb 16. iScience. 2024. PMID: 38327771 Free PMC article. - Whole-Exome Sequencing Identifies the VHL Mutation (c.262T > C, p.Try88Arg) in Non-Obstructive Azoospermia-Associated Cystic Renal Cell Carcinoma.
Man Y, Shang X, Liu C, Zhang W, Huang Q, Ma S, Shiang R, Zhang F, Zhang L, Zhang Z. Man Y, et al. Curr Oncol. 2022 Mar 28;29(4):2376-2384. doi: 10.3390/curroncol29040192. Curr Oncol. 2022. PMID: 35448166 Free PMC article. - Anti-CAIX BBζ CAR4/8 T cells exhibit superior efficacy in a ccRCC mouse model.
Wang Y, Buck A, Grimaud M, Culhane AC, Kodangattil S, Razimbaud C, Bonal DM, Nguyen QD, Zhu Z, Wei K, O'Donnell ML, Huang Y, Signoretti S, Choueiri TK, Freeman GJ, Zhu Q, Marasco WA. Wang Y, et al. Mol Ther Oncolytics. 2021 Dec 31;24:385-399. doi: 10.1016/j.omto.2021.12.019. eCollection 2022 Mar 17. Mol Ther Oncolytics. 2021. PMID: 35118195 Free PMC article. - Timing gone awry: distinct tumour suppressive and oncogenic roles of the circadian clock and crosstalk with hypoxia signalling in diverse malignancies.
Chang WH, Lai AG. Chang WH, et al. J Transl Med. 2019 Apr 23;17(1):132. doi: 10.1186/s12967-019-1880-9. J Transl Med. 2019. PMID: 31014368 Free PMC article.
References
- Maher E, Kaelin WG. von Hippel-Lindau Disease. Medicine. 1997;76:381–391. - PubMed
- Kim WY, Kaelin WG. Role of VHL Gene Mutation in Human Cancer. J Clin Onc. 2004;22:4991–5004. - PubMed
- Furge KA, Tan MH, Dykema K, Kort E, Stadler W, Yao X, et al. Identification of deregulated oncogenic pathways in renal cell carcinoma: an integrated oncogenomic approach based on gene expression profiling. Oncogene. 2007;26:1346–1350. - PubMed
- Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources