What's in a name? - PubMed (original) (raw)
Comment
What's in a name?
Mitchell J Weiss et al. J Clin Invest. 2012 Jul.
Abstract
Mutations in numerous genes encoding ribosomal proteins (RPs) occur in 50%-70% of individuals with Diamond-Blackfan anemia (DBA), establishing the disease as a ribosomopathy. As described in this issue of JCI, Sankaran, Gazda, and colleagues used genome-wide exome sequencing to study DBA patients with no detectable mutations in RP genes. They identified two unrelated pedigrees in which the disease is associated with mutations in GATA1, which encodes an essential hematopoietic transcription factor with no known mechanistic links to ribosomes. These findings ignite an interesting and potentially emotional debate on how we define DBA and whether the term should be restricted to pure ribosomopathies. More generally, the work reflects the powerful knowledge and controversies arising from the deluge of data generated by new genetic technologies that are being used to analyze human diseases.
Figures
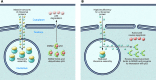
Figure 1. Current model for how RP haploinsufficiency causes DBA.
(A) Normal erythroblasts produce large numbers of ribosomes for protein synthesis. Levels of p53 remain low via a feedback loop whereby MDM2, a transcriptional p53 target, ubiquitinates p53 to promote its degradation by proteasomes. (B) Haploinsufficiency for specific RPs causes accumulation of other RPs, which bind to MDM2, thereby inhibiting its ability to promote p53 degradation. Consequently, p53 accumulates and triggers cell cycle arrest and apoptosis.
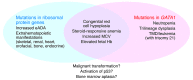
Figure 2. GATA1 mutations associated with human disease.
The diagram indicates GATA-1 protein with functional modules including the NH2-terminal activation domain (NAD), amino zinc finger (Nf), and carboxyl zinc finger (Cf). The NAD physically interacts with the retinoblastoma protein (Rb), which may modulate the ability of GATA-1 to regulate cell division and/or survival. Loss of the NAD through somatically acquired splice, frameshift, or nonsense mutations causes myeloproliferative disorder and leukemia in young children with Down syndrome (trisomy 21). In the absence of Down syndrome, germline mutations resulting in loss of the NAD are associated with congenital anemia. Different surfaces of the Nf interact with DNA (nor shown) and protein cofactors including FOG1 and SCL/TAL1. Missense mutations that alter these interaction surfaces of the Nf cause inherited anemia and/or thrombocytopenia with other abnormalities, as indicated.
Figure 3. Shared and distinct phenotypes in congenital red cell aplasia caused by mutations in RP genes and in GATA1.
eADA, erythrocyte adenosine deaminase activity, MCV, mean corpuscular volume; Hb, hemoglobin; TMD, transient myeloproliferative disorder.
Comment on
- Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia.
Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, Gazda HT. Sankaran VG, et al. J Clin Invest. 2012 Jul;122(7):2439-43. doi: 10.1172/JCI63597. Epub 2012 Jun 18. J Clin Invest. 2012. PMID: 22706301 Free PMC article.
Similar articles
- Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia.
Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, Gazda HT. Sankaran VG, et al. J Clin Invest. 2012 Jul;122(7):2439-43. doi: 10.1172/JCI63597. Epub 2012 Jun 18. J Clin Invest. 2012. PMID: 22706301 Free PMC article. - Altered translation of GATA1 in Diamond-Blackfan anemia.
Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, Lodish HF, Lander ES, Sankaran VG. Ludwig LS, et al. Nat Med. 2014 Jul;20(7):748-53. doi: 10.1038/nm.3557. Epub 2014 Jun 22. Nat Med. 2014. PMID: 24952648 Free PMC article. - Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28.
Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX; UW Center for Mendelian Genomics; Pilchman L, Sahraoui R, Stabley DL, Sol-Church K. Gripp KW, et al. Am J Med Genet A. 2014 Sep;164A(9):2240-9. doi: 10.1002/ajmg.a.36633. Epub 2014 Jun 18. Am J Med Genet A. 2014. PMID: 24942156 Free PMC article. - Ribosomopathies: how a common root can cause a tree of pathologies.
Danilova N, Gazda HT. Danilova N, et al. Dis Model Mech. 2015 Sep;8(9):1013-26. doi: 10.1242/dmm.020529. Dis Model Mech. 2015. PMID: 26398160 Free PMC article. Review. - Toward RNA Repair of Diamond Blackfan Anemia Hematopoietic Stem Cells.
D'Allard DL, Liu JM. D'Allard DL, et al. Hum Gene Ther. 2016 Oct;27(10):792-801. doi: 10.1089/hum.2016.081. Hum Gene Ther. 2016. PMID: 27550323 Review.
Cited by
- Molecular convergence in ex vivo models of Diamond-Blackfan anemia.
O'Brien KA, Farrar JE, Vlachos A, Anderson SM, Tsujiura CA, Lichtenberg J, Blanc L, Atsidaftos E, Elkahloun A, An X, Ellis SR, Lipton JM, Bodine DM. O'Brien KA, et al. Blood. 2017 Jun 8;129(23):3111-3120. doi: 10.1182/blood-2017-01-760462. Epub 2017 Apr 4. Blood. 2017. PMID: 28377399 Free PMC article. - [The progress of molecular genetics in bone marrow failure].
Liu CX, Zhang FK. Liu CX, et al. Zhonghua Xue Ye Xue Za Zhi. 2017 Jan 14;38(1):79-82. doi: 10.3760/cma.j.issn.0253-2727.2017.01.020. Zhonghua Xue Ye Xue Za Zhi. 2017. PMID: 28219235 Free PMC article. Chinese. No abstract available. - GATA factor mutations in hematologic disease.
Crispino JD, Horwitz MS. Crispino JD, et al. Blood. 2017 Apr 13;129(15):2103-2110. doi: 10.1182/blood-2016-09-687889. Epub 2017 Feb 8. Blood. 2017. PMID: 28179280 Free PMC article. Review. - Emerging cellular and gene therapies for congenital anemias.
Ludwig LS, Khajuria RK, Sankaran VG. Ludwig LS, et al. Am J Med Genet C Semin Med Genet. 2016 Dec;172(4):332-348. doi: 10.1002/ajmg.c.31529. Epub 2016 Oct 28. Am J Med Genet C Semin Med Genet. 2016. PMID: 27792859 Free PMC article. Review. - GATA1 and PU.1 Bind to Ribosomal Protein Genes in Erythroid Cells: Implications for Ribosomopathies.
Amanatiadou EP, Papadopoulos GL, Strouboulis J, Vizirianakis IS. Amanatiadou EP, et al. PLoS One. 2015 Oct 8;10(10):e0140077. doi: 10.1371/journal.pone.0140077. eCollection 2015. PLoS One. 2015. PMID: 26447946 Free PMC article.
References
- Diamond L, Blackfan K. Hypoplastic anemia. Am J Dis Child. 1938;56:464–467.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials