Substrate-specific transcription of the enigmatic GH61 family of the pathogenic white-rot fungus Heterobasidion irregulare during growth on lignocellulose - PubMed (original) (raw)
Substrate-specific transcription of the enigmatic GH61 family of the pathogenic white-rot fungus Heterobasidion irregulare during growth on lignocellulose
Igor Yakovlev et al. Appl Microbiol Biotechnol. 2012 Aug.
Abstract
The GH61 represents the most enigmatic Glycoside Hydrolase family (GH) regarding enzymatic activity and importance in cellulose degradation. Heterobasidion irregulare is a necrotizing pathogen and white-rot fungus that causes enormous damages in conifer forests. The genome of H. irregulare allowed identification of ten HiGH61 genes. qRT-PCR analysis separate the HiGH61 members into two groups; one that show up regulation on lignocellulosic substrates (HiGH61A, HiGH61B, HiGH61D, HiGH61G, HiGH61H, and HiGH61I) and a second showing either down-regulation or constitutive expression (HiGH61C, HiGH61E, HiGH61F, and HiGH61J). HiGH61H showed up to 17,000-fold increase on spruce heartwood suggesting a pivotal role in cellulose decomposition during saprotrophic growth. Sequence analysis of these genes reveals that all GH61s except HiGH61G possess the conserved metal-binding motif essential for activity. The sequences also divide into groups having either an insert near the N terminus or an insert near the second catalytic histidine, which may represent extensions of the substrate-binding surface. Three of the HiGH61s encode cellulose-binding modules (CBM1). Interestingly, HiGH61H and HiGH61I having CBM1s are up-regulated on pure cellulose. There was a common substrate-specific induction patterns of the HiGH61s with several reference cellulolytic and hemicellulolytic GHs, this taken together with their low transcript levels on media lacking lignocellulose, reflect the concerted nature of cell wall polymer degradation.
Figures
Fig. 1
Multiple sequence alignment of HiGH61 sequences and sequences of GH61 proteins with known 3D structures (TtGH61E from T. terrestris and HjGH61B from H. jecorina). Regions with insertions/deletions are shaded yellow (region 1) and blue (region 2). Conserved residues clustered on the putative substrate-binding surface/active site are shaded red. Degree of conservation: asterisks fully conserved, colons highly conserved, and full stops moderately conserved. Conserved residues with solvent exposed side chains (inferred by analysis of the HiGH61A 3D model by WHATIF) are marked with an “_e_”. CBM1 domains are indicated by a box and the conserved cysteines within these modules are colored green
Fig. 2
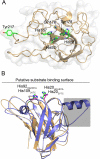
3D structural model of HiGH61A. a A superposition of the HiGH61A model (light-orange cartoon) and HjGH61B (PDB ID: 2VTC; blue cartoon) illustrating the binding surface extension seen in some of the HiGH61 proteins (sequence part covered by a grey shaded box; see also Fig. S1 in the ESM). The putative substrate-binding surface is seen from the side and also indicated by a line. The two conserved metal-binding histidines that are essential for GH61 activity (metal not shown) are shown in stick representation (colored green for HiGH61A and magenta for HjGH61B). b H. irregulare, a view looking down on the putative HiGH61A substrate-binding surface (i.e., rotated 90° around the _x_-axis compared with (a)) with the conserved, solvent-exposed amino acids shown in green stick representation. The protein is visualized in light-orange cartoon representation surrounded by a transparent white molecular surface
Fig. 3
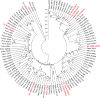
Phylogenetic tree of GH61 family proteins (full ORF amino acid sequences) available at the JGI fungal genomic database. Species abbreviation and protein ID are shown for each entry. The phylogenetic tree was constructed using MEGA5 as detailed in “Materials and methods.” The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Sequences from H. irregulare (Hir) are shown in red. Fungal species: Agabi A. bisporus, Pleos P. ostreatus, Pospl P. placenta, Lacbi L. bicolor, Phchr P. chrysosporium, Serla S. lacrymans, Schco S. commune, TtGH61E from T. terrestris and HjGH61B from H. jecorina
Fig. 4
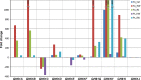
Transcript level profiles of GH61 polysaccharide oxidases of H. irregulare during fungal growth on different substrates from P. abies (Pa) and P. sylvestris (Ps). Substrates: Cel cellulose, RZ powdered reaction zone wood of Norway spruce, HW powdered heartwood wood of Norway spruce and Scots pine, SW powdered sapwood of Scots pine. Analysis was based on three biological replicates per culture medium. Transcript levels, normalized to the geometric mean of HiAct, HiαTub, and HiUBC2 Ct values (endogenous controls), are shown as fold changes in relation to their normalized transcript levels on the control medium (liquid Hagem)
Fig. 5
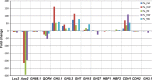
Transcript level profiles of selected lignocellulose active genes of H. irregulare (see legend of Fig. 4 for more information). Lcc laccase 2, Aao aryl-alcohol oxidase, QOr quinone oxidoreductase, СDH cellobiose dehydrogenase, HBF hydrophobin, GH glycoside hydrolase
Similar articles
- Genes associated with lignin degradation in the polyphagous white-rot pathogen Heterobasidion irregulare show substrate-specific regulation.
Yakovlev IA, Hietala AM, Courty PE, Lundell T, Solheim H, Fossdal CG. Yakovlev IA, et al. Fungal Genet Biol. 2013 Jul;56:17-24. doi: 10.1016/j.fgb.2013.04.011. Epub 2013 May 9. Fungal Genet Biol. 2013. PMID: 23665189 - Biochemical studies of two lytic polysaccharide monooxygenases from the white-rot fungus Heterobasidion irregulare and their roles in lignocellulose degradation.
Liu B, Olson Å, Wu M, Broberg A, Sandgren M. Liu B, et al. PLoS One. 2017 Dec 11;12(12):e0189479. doi: 10.1371/journal.pone.0189479. eCollection 2017. PLoS One. 2017. PMID: 29228039 Free PMC article. - Xylem defense wood of Norway spruce compromised by the pathogenic white-rot fungus Heterobasidion parviporum shows a prolonged period of selective decay.
Nagy NE, Ballance S, Kvaalen H, Fossdal CG, Solheim H, Hietala AM. Nagy NE, et al. Planta. 2012 Oct;236(4):1125-33. doi: 10.1007/s00425-012-1664-4. Epub 2012 May 27. Planta. 2012. PMID: 22644766 - Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium.
Gold MH, Alic M. Gold MH, et al. Microbiol Rev. 1993 Sep;57(3):605-22. doi: 10.1128/mr.57.3.605-622.1993. Microbiol Rev. 1993. PMID: 8246842 Free PMC article. Review. - Phanerochaete chrysosporium and its natural substrate.
Broda P, Birch P, Brooks P, Copa-Patiño JL, Sinnott ML, Tempelaars C, Wang Q, Wyatt A, Sims P. Broda P, et al. FEMS Microbiol Rev. 1994 Mar;13(2-3):189-95. doi: 10.1111/j.1574-6976.1994.tb00042.x. FEMS Microbiol Rev. 1994. PMID: 8167034 Review.
Cited by
- The copper active site of CBM33 polysaccharide oxygenases.
Hemsworth GR, Taylor EJ, Kim RQ, Gregory RC, Lewis SJ, Turkenburg JP, Parkin A, Davies GJ, Walton PH. Hemsworth GR, et al. J Am Chem Soc. 2013 Apr 24;135(16):6069-77. doi: 10.1021/ja402106e. Epub 2013 Apr 10. J Am Chem Soc. 2013. PMID: 23540833 Free PMC article. - Distinct Substrate Specificities and Electron-Donating Systems of Fungal Lytic Polysaccharide Monooxygenases.
Frommhagen M, Westphal AH, van Berkel WJH, Kabel MA. Frommhagen M, et al. Front Microbiol. 2018 May 29;9:1080. doi: 10.3389/fmicb.2018.01080. eCollection 2018. Front Microbiol. 2018. PMID: 29896168 Free PMC article. Review. - Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio japonicus.
Forsberg Z, Nelson CE, Dalhus B, Mekasha S, Loose JS, Crouch LI, Røhr ÅK, Gardner JG, Eijsink VG, Vaaje-Kolstad G. Forsberg Z, et al. J Biol Chem. 2016 Apr 1;291(14):7300-12. doi: 10.1074/jbc.M115.700161. Epub 2016 Feb 8. J Biol Chem. 2016. PMID: 26858252 Free PMC article. - Function-based classification of carbohydrate-active enzymes by recognition of short, conserved peptide motifs.
Busk PK, Lange L. Busk PK, et al. Appl Environ Microbiol. 2013 Jun;79(11):3380-91. doi: 10.1128/AEM.03803-12. Epub 2013 Mar 22. Appl Environ Microbiol. 2013. PMID: 23524681 Free PMC article. - Plant-polysaccharide-degrading enzymes from Basidiomycetes.
Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. Rytioja J, et al. Microbiol Mol Biol Rev. 2014 Dec;78(4):614-49. doi: 10.1128/MMBR.00035-14. Microbiol Mol Biol Rev. 2014. PMID: 25428937 Free PMC article. Review.
References
- DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002.
- Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee Y-H, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie X, Kües U, Hibbett DS, Hoffmeister D, Högberg N, Martin F, Grigoriev IV, Watkinson SC. The plant cell wall–decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources