Functional characterization of the role of the N-terminal domain of the c/Nip1 subunit of eukaryotic initiation factor 3 (eIF3) in AUG recognition - PubMed (original) (raw)
Functional characterization of the role of the N-terminal domain of the c/Nip1 subunit of eukaryotic initiation factor 3 (eIF3) in AUG recognition
Martina Karásková et al. J Biol Chem. 2012.
Abstract
In eukaryotes, for a protein to be synthesized, the 40 S subunit has to first scan the 5'-UTR of the mRNA until it has encountered the AUG start codon. Several initiation factors that ensure high fidelity of AUG recognition were identified previously, including eIF1A, eIF1, eIF2, and eIF5. In addition, eIF3 was proposed to coordinate their functions in this process as well as to promote their initial binding to 40 S subunits. Here we subjected several previously identified segments of the N-terminal domain (NTD) of the eIF3c/Nip1 subunit, which mediates eIF3 binding to eIF1 and eIF5, to semirandom mutagenesis to investigate the molecular mechanism of eIF3 involvement in these reactions. Three major classes of mutant substitutions or internal deletions were isolated that affect either the assembly of preinitiation complexes (PICs), scanning for AUG, or both. We show that eIF5 binds to the extreme c/Nip1-NTD (residues 1-45) and that impairing this interaction predominantly affects the PIC formation. eIF1 interacts with the region (60-137) that immediately follows, and altering this contact deregulates AUG recognition. Together, our data indicate that binding of eIF1 to the c/Nip1-NTD is equally important for its initial recruitment to PICs and for its proper functioning in selecting the translational start site.
Figures
FIGURE 1.
A three-dimensional model of eIF3 and its associated eIFs in the MFC (adapted from Ref. 13). ntd, N-terminal domain; ctd, C-terminal domain; hld, Hcr1-like domain; rrm, RNA recognition motif; pci, PCI domain. The NMR structure of the interaction between the RNA recognition motif of human eIF3b (green and light blue) and the N-terminal peptide of human eIF3j (yellow) (9), the NMR structure of the C-terminal RNA recognition motif of human eIF3g (red and sky blue) (18), the x-ray structure of the yeast i/Tif34·b/Prt1-CTD complex (13), and the three-dimensional homology model of the c/Nip1-CTD (31) were used to replace the original schematic representations of the corresponding molecules.
FIGURE 2.
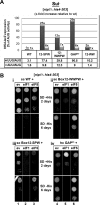
Semirandom mutagenesis of the preselected segments (Boxes) of the extreme NTD of c/Nip1 yields mutants displaying either Sui− or Gcd− phenotypes alone or in combination. A, amino acid sequence alignment of the Boxes of interest from the c/Nip1-NTD of Saccharomyces cerevisiae with that of other species. The amino acid sequence of S. cerevisiae c/Nip1 (accession number P32497.2 UniProtKB/Swiss-Prot) between residues 1 and 160 was aligned with its Caenorhabditis elegans (accession number A8WWU0.1 UniProtKB/Swiss-Prot), Homo sapiens (accession number Q99613.1 UniProtKB/Swiss-Prot), Arabidopsis thaliana (accession number AAC83464.1 GenBank), and Schizosaccharomyces pombe (accession number CAB11485.2 GenBank) homologues using ClustalW; only the sequences corresponding to the Boxes of interest are shown. Highly conserved residues are color-coded. Altogether, nine mutant libraries containing random amino acid residues at selected, color-coded sites marked with “_X_” and grouped together individually for each Box are indicated. B, schematic representation of the first 160 amino acid residues of c/Nip1 shown as numbered circles (Boxes 1–16), each of which is composed of 10 consecutive residues. Three Boxes of interest are color-coded, and the sequences and phenotypes of the mutants derived from the semirandom mutagenesis are given below the schematic. Two internal deletions that were unintentionally generated by this mutagenic procedure are also shown with the deleted residues indicated. A table to the right of the schematic summarizes phenotypes associated with individual mutations including Slg−, Sui−, Gcd−, and the polysome/monosome (P/M) ratios (averaged values of replicate experiments are given; S.E. values ranged between 5 and 15% and are not shown). C, the c/Nip1 mutants producing the Sui− phenotype indicative of a defect in AUG recognition. The HLV04 (_nip1_Δ his4-303) strain was transformed with the corresponding sc or hc plasmids carrying WT NIP1 and its mutant alleles, and the resident pNIP1+ (NIP1, URA3) covering plasmid was evicted on 5-fluoro-orotic acid. The resulting strains and the parental control strain TD301-8D (NIP1 sui1-1 his4-303) were then spotted in four serial 10-fold dilutions on SD medium containing histidine (upper panel) or lacking histidine (lower panel) and incubated at 30 °C for 2 (upper panel) or 7 days (lower panel). D, the c/Nip1 mutants producing the Gcd− phenotype indicative of a defect in the assembly of the PICs. The HKN06 (_nip1_Δ _gcn2_Δ) strain was transformed with the corresponding sc or hc plasmids carrying WT NIP1 and its mutant alleles, and the resident pNIP1+ (NIP1, URA3) covering plasmid was evicted on 5-fluoro-orotic acid. The resulting transformants and isogenic strains H2880 (GCN2) (lane 1) and H2881 (_gcn2_Δ) (lane 2) transformed with empty vector were spotted in four serial dilutions on SD (upper panel) or SD medium containing 30 m
m
3-AT (lower panel) and incubated at 30 °C for 3 and 6 days, respectively. E, Western blot analysis of the WCEs derived from the indicated strains described in C using antibodies raised against c/Nip1 and Rps0A.
FIGURE 3.
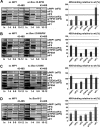
High dosage suppressor analysis of the c/Nip1-NTD Sui− mutants by the selected eIFs known to participate in the AUG recognition process. A, quantification of Sui− phenotypes. Strains from Fig. 2_C_ were transformed with HIS4-lacZ reporters with AUG (p367), UUG (p391), or AUU (p2042), respectively, and grown in SD medium supplemented with His and Trp at 30 °C, and β-galactosidase activities were measured in WCEs. The mean percentages of the UUG or AUU initiation rates relative to those of AUG and an x_-fold increase over the WT c/Nip1 were calculated from three experiments with six independent transformants. B, genetic effects of high copy expression of the indicated eIFs on selected Sui− mutants of c/Nip1. Strains from Fig. 2_C were transformed with either empty vector (ev; lanes 1 and 4) or the corresponding plasmids overexpressing SUI1 (eIF1; lanes 2 and 5) or TIF5 (eIF5; lanes 3 and 6), respectively, and the resulting transformants were spotted in three serial dilutions on SD medium containing histidine (upper panels i and ii) or lacking histidine (lower panels iii and iv) and incubated at 30 °C for 2 (upper panels i and ii) or 6 days (lower panels iii and iv).
FIGURE 4.
The c/nip1-Boxes 12-WWPW, 12-WW, and 15-2 impair GCN4 translational control (produce the Gcd− phenotype) in the canonical manner suppressible by overexpressing the TC. A, strains from Fig. 2_D_ were transformed with either empty vector or all three subunits of eIF2 + IMT4 (TC) on a high copy plasmid. The resulting transformants and isogenic strains H2880 (GCN2) and H2881 (_gcn2_Δ) transformed with empty vector were spotted in four serial dilutions on SD medium (upper panel) or SD medium containing 30 m
m
3-AT (lower panel) and incubated at 30 °C for 2 and 7 days, respectively. B, quantification of the Gcd− phenotypes with or without the hc TC effect. Selected _nip1_Δ strains from A were further transformed with p180 containing the GCN4-lacZ fusion with all four uORFs present (shown as a schematic above the plot) and grown in minimal medium for 6 h, and the β-galactosidase activities were measured in the WCEs and expressed in units of nmol of _o-_nitrophenyl-β-
d
-galactopyranoside hydrolyzed/min/mg of protein. The mean values and standard deviations obtained from at least six independent measurements with three independent transformants, the _x_-fold increases in activities of the mutant strains relative to the corresponding WT expressing either empty vector (EV) or hc TC, and percentages of hc TC activities relative to empty vector activities in each individual strain (indicating the extent of the hc TC suppression effect) are given in the histogram. nd, not done.
FIGURE 5.
The extreme N terminus of c/Nip1 interacts with eIF5, the segment that immediately follows binds eIF1, and nip1-Box12-SPW increases binding affinity of the c/Nip1-NTD for eIF1 in vitro. A, schematic as in Fig. 2_B_ showing the minimal binding segments (as arrows) of the c/Nip1-NTD for eIFs 1 and 5. The lines beneath the arrows depict the 35S-labeled segments of the c/Nip1-NTD used in the binding assays of B with the amino acid end points and clone designations indicated. The binding of each c/Nip1 construct to GST-eIF1 or -eIF5 is summarized in the table on the right. Data from the WT ((N)*) construct were taken from Ref. for comparison purposes only. B, eIF1 and eIF5 fused to GST (lanes 3 and 4) or GST alone (lane 2) was expressed in E. coli, immobilized on glutathione-Sepharose beads, and incubated with the 35S-labeled c/Nip1 segments indicated to the right of the lower panels at 4 °C for 2 h. The beads were washed three times with phosphate-buffered saline, and bound proteins were separated by SDS-PAGE. Gels were first stained with GelCode Blue Stain Reagent (Pierce) (top panel) followed by autoradiography (bottom panels). Lane 1 shows 10% of the input (In) amounts. C, same as in B except that the semirandom mutations of c/Nip1-Boxes are under study. The amount of each 35S-labeled c/Nip1 polypeptide bound to each GST fusion protein was quantified and is expressed above the corresponding panel as a percentage of the input (three independent measurements were made, and the outputs were averaged with the S.E. ranging between 3 and 11%).
FIGURE 6.
The semirandom mutations of c/Nip1-Boxes selectively affect composition of the MFC in vivo. A, WCEs prepared from the selected strains described in Fig. 2_C_ were incubated with Ni2+-Sepharose, and the bound proteins were eluted and subjected to Western blot analysis with the antibodies indicated in each row. In, lanes contained 5% of the input WCEs; E, lanes contained 100% of the eluate from the resin (a typical recovery is ∼10% of the input for the WT); FT, lanes contained 5% of the flow-through from 100% input that did not bind to the resin (the limiting factor in this experiment). B, the Western signals for the indicated proteins in the eluate fractions of the WT NIP1-His and its mutants were quantified, normalized for the amounts of the WT c/Nip1 in these fractions, and plotted in the histogram as percentages of the corresponding values calculated for the WT c/Nip1; S.D. values are given.
FIGURE 7.
The c/Nip1-NTD mutants generally reduce 40 S subunit association of eIF3 and eIF5 in vivo but have varying effects on eIF1 and TC recruitment to the 43–48 S PICs. A–D, selected strains described in Fig. 2, C and D, were grown in YPD medium at 30 °C to an _A_600 of ∼1.5 and cross-linked with 2% HCHO prior to harvesting. WCEs were prepared, separated on a 7.5–30% sucrose gradient by centrifugation at 41,000 rpm for 5 h, and subjected to Western blot analysis. Fractions 1–4, 5–9, and 10–12 (43–48 S) were pooled, and 5% of each pooled sample was loaded on the gel; lane “_In_” shows 5% input. Proportions of the 40 S subunit-bound proteins relative to the amount of 40 S subunits were calculated using Quantity One software (Bio-Rad) from at least three independent experiments. The resulting values obtained with the WT strain were set to 100%, and those obtained with mutant strains were expressed as percentages of the WT in the histograms on the right (S.D. values are given).
Similar articles
- Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection.
Valásek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Valásek L, et al. Mol Cell Biol. 2004 Nov;24(21):9437-55. doi: 10.1128/MCB.24.21.9437-9455.2004. Mol Cell Biol. 2004. PMID: 15485912 Free PMC article. - β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo.
Martin-Marcos P, Nanda J, Luna RE, Wagner G, Lorsch JR, Hinnebusch AG. Martin-Marcos P, et al. J Biol Chem. 2013 Sep 20;288(38):27546-27562. doi: 10.1074/jbc.M113.498642. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893413 Free PMC article. - The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes.
Kouba T, Rutkai E, Karásková M, Valášek L. Kouba T, et al. Nucleic Acids Res. 2012 Mar;40(6):2683-99. doi: 10.1093/nar/gkr1083. Epub 2011 Nov 28. Nucleic Acids Res. 2012. PMID: 22123745 Free PMC article. - The scanning mechanism of eukaryotic translation initiation.
Hinnebusch AG. Hinnebusch AG. Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review. - Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation.
Hinnebusch AG. Hinnebusch AG. Trends Biochem Sci. 2017 Aug;42(8):589-611. doi: 10.1016/j.tibs.2017.03.004. Epub 2017 Apr 22. Trends Biochem Sci. 2017. PMID: 28442192 Review.
Cited by
- Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex.
Aitken CE, Beznosková P, Vlčkova V, Chiu WL, Zhou F, Valášek LS, Hinnebusch AG, Lorsch JR. Aitken CE, et al. Elife. 2016 Oct 26;5:e20934. doi: 10.7554/eLife.20934. Elife. 2016. PMID: 27782884 Free PMC article. - Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Bochler A, Querido JB, Prilepskaja T, Soufari H, Simonetti A, Del Cistia ML, Kuhn L, Ribeiro AR, Valášek LS, Hashem Y. Bochler A, et al. Cell Rep. 2020 Dec 22;33(12):108534. doi: 10.1016/j.celrep.2020.108534. Cell Rep. 2020. PMID: 33357443 Free PMC article. - Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle.
Valášek LS, Zeman J, Wagner S, Beznosková P, Pavlíková Z, Mohammad MP, Hronová V, Herrmannová A, Hashem Y, Gunišová S. Valášek LS, et al. Nucleic Acids Res. 2017 Nov 2;45(19):10948-10968. doi: 10.1093/nar/gkx805. Nucleic Acids Res. 2017. PMID: 28981723 Free PMC article. - Lithium Chloride Sensitivity in Yeast and Regulation of Translation.
Hajikarimlou M, Hunt K, Kirby G, Takallou S, Jagadeesan SK, Omidi K, Hooshyar M, Burnside D, Moteshareie H, Babu M, Smith M, Holcik M, Samanfar B, Golshani A. Hajikarimlou M, et al. Int J Mol Sci. 2020 Aug 10;21(16):5730. doi: 10.3390/ijms21165730. Int J Mol Sci. 2020. PMID: 32785068 Free PMC article. - The eIF3 complex of Trypanosoma brucei: composition conservation does not imply the conservation of structural assembly and subunits function.
Li K, Zhou S, Guo Q, Chen X, Lai DH, Lun ZR, Guo X. Li K, et al. RNA. 2017 Mar;23(3):333-345. doi: 10.1261/rna.058651.116. Epub 2016 Dec 8. RNA. 2017. PMID: 27932584 Free PMC article.
References
- Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26, 41–50 - PubMed
- Algire M. A., Maag D., Lorsch J. R. (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20, 251–262 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous