Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition - PubMed (original) (raw)
doi: 10.1371/journal.pone.0026284. Epub 2012 Jun 13.
Christina M Hamm, Ajay S Gulati, R Balfour Sartor, Hongyan Chen, Xiao Wu, Tianyi Zhang, F James Rohlf, Wei Zhu, Chi Gu, Charles E Robertson, Norman R Pace, Edgar C Boedeker, Noam Harpaz, Jeffrey Yuan, George M Weinstock, Erica Sodergren, Daniel N Frank
Affiliations
- PMID: 22719818
- PMCID: PMC3374607
- DOI: 10.1371/journal.pone.0026284
Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition
Ellen Li et al. PLoS One. 2012.
Abstract
We tested the hypothesis that Crohn's disease (CD)-related genetic polymorphisms involved in host innate immunity are associated with shifts in human ileum-associated microbial composition in a cross-sectional analysis of human ileal samples. Sanger sequencing of the bacterial 16S ribosomal RNA (rRNA) gene and 454 sequencing of 16S rRNA gene hypervariable regions (V1-V3 and V3-V5), were conducted on macroscopically disease-unaffected ileal biopsies collected from 52 ileal CD, 58 ulcerative colitis and 60 control patients without inflammatory bowel diseases (IBD) undergoing initial surgical resection. These subjects also were genotyped for the three major NOD2 risk alleles (Leu1007fs, R708W, G908R) and the ATG16L1 risk allele (T300A). The samples were linked to clinical metadata, including body mass index, smoking status and Clostridia difficile infection. The sequences were classified into seven phyla/subphyla categories using the Naïve Bayesian Classifier of the Ribosome Database Project. Centered log ratio transformation of six predominant categories was included as the dependent variable in the permutation based MANCOVA for the overall composition with stepwise variable selection. Polymerase chain reaction (PCR) assays were conducted to measure the relative frequencies of the Clostridium coccoides - Eubacterium rectales group and the Faecalibacterium prausnitzii spp. Empiric logit transformations of the relative frequencies of these two microbial groups were included in permutation-based ANCOVA. Regardless of sequencing method, IBD phenotype, Clostridia difficile and NOD2 genotype were selected as associated (FDR ≤ 0.05) with shifts in overall microbial composition. IBD phenotype and NOD2 genotype were also selected as associated with shifts in the relative frequency of the C. coccoides--E. rectales group. IBD phenotype, smoking and IBD medications were selected as associated with shifts in the relative frequency of F. prausnitzii spp. These results indicate that the effects of genetic and environmental factors on IBD are mediated at least in part by the enteric microbiota.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Phyla/subphyla comparison of three disease phenotypes (ileal CD, colitis, using the Sanger, 454 V1–V3 and 454 V3–V5 data sets.
The average relative frequency of each taxa is shown for ileal CD, colitis and control subjects for each of the three sequencing data sets, (see also Table S1 for means ± standard deviations).
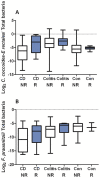
Figure 2. Targeted qPCR results for the C. coccoides-E. rectales group and for F. prausnitzii spp.
Boxplots of (panel A) the log2 C. coccoides-E. rectales group/total bacteria and (panel B) the log2 F. prausnitzii/total bacteria as a function of disease phenotype and NOD2 genotype assayed using qPCR are shown. The middle line represents the median, and the lower edge and the upper edge of the box represent the 25% and 75% quartiles. The bottom and top lines represent the minimum and maximum values, respectively. For the C. coccoides-rectales group, all 170 samples were assayed. For F. prausnitzii, 157 of 170 samples were assayed.
References
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. - PubMed
- Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect. Dis. 2007;454:256–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 HG005964/HG/NHGRI NIH HHS/United States
- NIH R21HG005964/HG/NHGRI NIH HHS/United States
- P30 DK52574/DK/NIDDK NIH HHS/United States
- UH2 DK083994/DK/NIDDK NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UH2DK083994/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous