Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains - PubMed (original) (raw)
Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains
Kevin Van Bortle et al. Genome Res. 2012 Nov.
Abstract
Several multiprotein DNA complexes capable of insulator activity have been identified in Drosophila melanogaster, yet only CTCF, a highly conserved zinc finger protein, and the transcription factor TFIIIC have been shown to function in mammals. CTCF is involved in diverse nuclear activities, and recent studies suggest that the proteins with which it associates and the DNA sequences that it targets may underlie these various roles. Here we show that the Drosophila homolog of CTCF (dCTCF) aligns in the genome with other Drosophila insulator proteins such as Suppressor of Hairy wing [SU(HW)] and Boundary Element Associated Factor of 32 kDa (BEAF-32) at the borders of H3K27me3 domains, which are also enriched for associated insulator proteins and additional cofactors. RNAi depletion of dCTCF and combinatorial knockdown of gene expression for other Drosophila insulator proteins leads to a reduction in H3K27me3 levels within repressed domains, suggesting that insulators are important for the maintenance of appropriate repressive chromatin structure in Polycomb (Pc) domains. These results shed new insights into the roles of insulators in chromatin domain organization and support recent models suggesting that insulators underlie interactions important for Pc-mediated repression. We reveal an important relationship between dCTCF and other Drosophila insulator proteins and speculate that vertebrate CTCF may also align with other nuclear proteins to accomplish similar functions.
Figures
Figure 1.
dCTCF aligns with Drosophila insulator-binding proteins SU(HW) and BEAF-32. (A) Venn diagram depicting overlap of dCTCF binding sites with those of BEAF-32 and SU(HW). Overlap represented as number of sites (summits ±150 bp) in which dCTCF intersects BEAF-32 and/or SU(HW) and target sequences are present for each insulator protein, suggesting close alignment (within 150 bp). (B) Example of ChIP-seq profile for dCTCF, BEAF-32, SU(HW), and CP190 on chromosome 3L; in which case dCTCF aligns with both BEAF-32 and SU(HW), where each cognate target is present. (C) Number of sites in which dCTCF, BEAF-32, and SU(HW) contain appropriate target sequences. Percentages of sites in which dCTCF, BEAF-32, and SU(HW) align closely with other DNA-binding insulator proteins. Most (90%) alignments include dCTCF, and as many as 40% of dCTCF sites align with either BEAF-32 and/or SU(HW).
Figure 2.
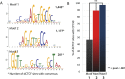
dCTCF sites are enriched for three distinct DNA motifs, including a similar but novel secondary motif enriched for insulator protein CP190. (A) Position weight matrices for primary target sequence and secondary target sequences obtained by MEME-ChIP, and confirmed with Weeder 1.3. Number of sites provided represents sites in which dCTCF summits are ±150 bp from the DNA motif. (B) Percentage of dCTCF sites in which CP190 is present when containing each DNA motif.
Figure 3.
dCTCF and BEAF-32 recruit isoform(s) of MOD(MDG4) different from MOD(MDG4)2.2. Aligned dCTCF sites are enriched for MOD(MDG4) and additional cofactors. (A) Immunofluorescence microscopy of MOD(MDG4) (green) and MOD(MDG4)2.2 (red) on Drosophila polytene chromosomes. MOD(MDG4) staining includes many discrete bands not accounted for by MOD(MDG4)2.2 specific antibodies, depicted with white arrows. (B) Genome-wide overlap of dCTCF, BEAF-32, and SU(HW) with MOD(MDG4) and MOD(MDG4)2.2. Many dCTCF (45%) and BEAF-32 (34%) sites overlap MOD(MDG4) isoform(s) not represented by MOD(MDG4)2.2. Meanwhile, many SU(HW) (37%) sites overlap MOD(MDG4)2.2 sites, as expected. (C) ChIP-seq profile for MOD(MDG4) and MOD(MDG4)2.2 reveals many unique peaks specifically in the MOD(MDG4) profile, accounted for at BEAF-32 and dCTCF sites. (D–F) Heatmap representation of percentages of dCTCF sites in which CP190, MOD(MDG4), MOD(MDG4)2.2, BEAF-32, SU(HW), L(3)MBT, and/or Chromator co-occur at independent dCTCF sites, aligned dCTCF sites, and sites where dCTCF aligns with both BEAF-32 and SU(HW).
Figure 4.
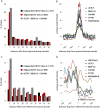
Aligned dCTCF sites are enriched at the borders of H3K27me3 and physical domains. (A) Percentage of independent and aligned dCTCF sites within 5 kb from recently mapped physical domain boundaries (Sexton et al. 2012). (B) Average read intensity for insulator proteins at physical domain boundaries, ±10 kb. Comparison of insulator profiles normalized by total read numbers. (C) Percentage of independent and aligned dCTCF sites within 5 kb from H3K27me3 domain borders. (D) Average read intensity for H3K27me3 and insulator proteins at H3K27me3 domain borders, ±2 kb. Comparison of insulator profiles normalized by total read numbers.
Figure 5.
RNAi depletion of insulator proteins causes H3K27me3 depletion within domains but has no effect on H3K27me3 domain flanking genes. (A) Percentage of genes down-regulated (more than twofold) in Drosophila Kc cells after knockdown of insulator protein expression. Comparison of genes flanking H3K27me3 domains with genome-wide averages. (B) Average read intensity for H3K27me3 in Drosophila Kc cells at domain borders and surrounding domain centers. H3K27me3 levels were determined by ChIP-seq before and after dCTCF knockdown. For appropriate comparison, ChIP-seq data for H3K27me3 was rank normalized as previously described (Whyte et al. 2012) and represented as average read intensity.
Figure 6.
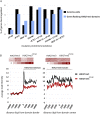
Insulator proteins, including MOD(MDG4), are necessary for the maintenance of H3K27me3 levels at several loci. (A–C) ChIP-seq profiles for insulator proteins and H3K27me3 levels before and after dCTCF knockdown at the even skipped, eyes absent, and Ods-site homeobox gene loci, respectively. (D–F) ChIP–PCR levels determined by qRT–PCR against H3K27me3 ChIP before and after knockdown of several insulator proteins. Control sample is represented as SEM from three independent biological replicates.
Figure 7.
Diagram comparing independent dCTCF sites and dCTCF sites aligned with BEAF-32 and SU(HW). dCTCF, BEAF-32, and SU(HW) function similarly through the recruitment of CP190 and MOD(MDG4). Aligned dCTCF sites are enriched for the secondary DNA sequence and CP190. Ultimately, alignment may allow for cooperative recruitment of CP190 and MOD(MDG4), ensuring that dCTCF establishes a functional insulator complex at domain borders. Recruitment of additional proteins, such as L(3)MBT and Chromator, may also contribute to insulator activities at these loci.
Similar articles
- Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila.
Gerasimova TI, Lei EP, Bushey AM, Corces VG. Gerasimova TI, et al. Mol Cell. 2007 Dec 14;28(5):761-72. doi: 10.1016/j.molcel.2007.09.024. Mol Cell. 2007. PMID: 18082602 Free PMC article. - The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions.
Oliver D, Sheehan B, South H, Akbari O, Pai CY. Oliver D, et al. BMC Cell Biol. 2010 Dec 31;11:101. doi: 10.1186/1471-2121-11-101. BMC Cell Biol. 2010. PMID: 21194420 Free PMC article. - The phylogenetic distribution of non-CTCF insulator proteins is limited to insects and reveals that BEAF-32 is Drosophila lineage specific.
Schoborg TA, Labrador M. Schoborg TA, et al. J Mol Evol. 2010 Jan;70(1):74-84. doi: 10.1007/s00239-009-9310-x. Epub 2009 Dec 19. J Mol Evol. 2010. PMID: 20024537 - Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.
Ahanger SH, Shouche YS, Mishra RK. Ahanger SH, et al. Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review. - Chromatin insulators: regulatory mechanisms and epigenetic inheritance.
Bushey AM, Dorman ER, Corces VG. Bushey AM, et al. Mol Cell. 2008 Oct 10;32(1):1-9. doi: 10.1016/j.molcel.2008.08.017. Mol Cell. 2008. PMID: 18851828 Free PMC article. Review.
Cited by
- Nuclear organization and genome function.
Van Bortle K, Corces VG. Van Bortle K, et al. Annu Rev Cell Dev Biol. 2012;28:163-87. doi: 10.1146/annurev-cellbio-101011-155824. Epub 2012 Aug 17. Annu Rev Cell Dev Biol. 2012. PMID: 22905954 Free PMC article. Review. - Insulator function and topological domain border strength scale with architectural protein occupancy.
Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. Van Bortle K, et al. Genome Biol. 2014 Jun 30;15(6):R82. doi: 10.1186/gb-2014-15-5-r82. Genome Biol. 2014. PMID: 24981874 Free PMC article. - Chromatin accessibility-based characterisation of brain gene regulatory networks in three distinct honey bee polyphenisms.
Lowe R, Wojciechowski M, Ellis N, Hurd PJ. Lowe R, et al. Nucleic Acids Res. 2022 Nov 11;50(20):11550-11562. doi: 10.1093/nar/gkac992. Nucleic Acids Res. 2022. PMID: 36330958 Free PMC article. - Spectral identification of topological domains.
Chen J, Hero AO 3rd, Rajapakse I. Chen J, et al. Bioinformatics. 2016 Jul 15;32(14):2151-8. doi: 10.1093/bioinformatics/btw221. Epub 2016 May 5. Bioinformatics. 2016. PMID: 27153657 Free PMC article. - Characterization of Button Loci that Promote Homologous Chromosome Pairing and Cell-Type-Specific Interchromosomal Gene Regulation.
Viets K, Sauria MEG, Chernoff C, Rodriguez Viales R, Echterling M, Anderson C, Tran S, Dove A, Goyal R, Voortman L, Gordus A, Furlong EEM, Taylor J, Johnston RJ Jr. Viets K, et al. Dev Cell. 2019 Nov 4;51(3):341-356.e7. doi: 10.1016/j.devcel.2019.09.007. Epub 2019 Oct 10. Dev Cell. 2019. PMID: 31607649 Free PMC article.
References
- Armknecht S, Boutros M, Kiger A, Nybakken K, Mathey-Prevot B, Perrimon N 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol 392: 55–73 - PubMed
- Bonchuk A, Denisov S, Georgiev P, Maksimenko O 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J Mol Biol 412: 423–436 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases