Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation - PubMed (original) (raw)
Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation
Celeste Holz-Schietinger et al. J Biol Chem. 2012.
Abstract
DNA methylation is a key regulator of gene expression and changes in DNA methylation occur early in tumorigenesis. Mutations in the de novo DNA methyltransferase gene, DNMT3A, frequently occur in adult acute myeloid leukemia patients with poor prognoses. Most of the mutations occur within the dimer or tetramer interface, including Arg-882. We have identified that the most prevalent mutation, R882H, and three additional mutants along the tetramer interface disrupt tetramerization. The processive methylation of multiple CpG sites is disrupted when tetramerization is eliminated. Our results provide a possible mechanism that accounts for how DNMT3A mutations may contribute to oncogenesis and its progression.
Figures
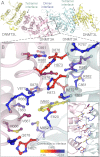
FIGURE 1.
DNMT3A homodimer interface. A, DNMT3A-DNMT3L heterotetramer based on the crystal structure (Protein Data Bank code 2QRV), showing the dimer and tetramer interface; the boxed region is enlarged below. B, the dimer interface is symmetrical; shown is a close-up of half the interface and all of the center interactions. Underlined residues were mutated in this study. The interface has many ionic interactions and a few aromatic residues. Residues are colored based upon their predicted energetic contribution to the dimer interface, in ΔΔ_G_, compared with alanine as determined using the Rosetta interface alanine scanning module. Bright yellow residues provide the greatest contribution to the DNMT3A-DNMT3A interface. Boxed is the full dimer interface showing both symmetrical sides of the dimer interface.
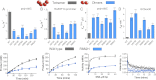
FIGURE 2.
Concentration-dependent DNMT3A activation. A, the activity of DNMT3A shows a sigmoidal relationship with protein concentration. DNMT3A is more active at higher enzyme concentrations suggesting that DNMT3A oligomerization is concentration-dependent. The DNA is below the Km at 500 n
m
bp. B, DNMT3A with DNMT3L is more active at higher DNMT3A concentrations (DNMT3L is kept constant at saturating concentrations), suggesting that the concentration-dependent oligomerization takes place at the dimer interface (the interface that does not bind DNMT3L). The DNA is below the Km at 500 n
m
bp. C, the activity of DNMT3A shows a linear relationship when DNA is at over above the Km, data shown are when DNA is at 2 μ
m
bp. Reactions took place on poly-dIdC, a multiple site substrate.
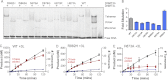
FIGURE 3.
Dimer interface mutants disrupt the interface resulting in dimers on DNA. A, size-exclusion chromatography of light scattering traces of tetrameric wild type catalytic domain (black trace) and representative dimeric H873A (blue trace). Molecular weights were determined from the amount of scattered light, in relation to protein concentration determined by _A_280. B, diagram of oligomeric mutants with and without DNA. C, EMSA of size markers, DNA (GCbox30) has binding sites for size standards, one site for M. HhaI (37 kDa), a known monomer and two binding sites for EcoRV (29 kDa), a known dimer, which creates dimer and tetramer bands. D, EMSA showed mutations at the dimer interface of DNMT3A disrupt oligomerization resulting in dimers on DNA (200 n
m
enzyme, 300 n
m
DNA). E, the wild type DNMT3A at varying concentrations (20–500 n
m
) migrates as a tetramer or larger on DNA (200 n
m
). F, the H873A mutant (20–500 n
m
) migrates as a dimer on DNA (200 n
m
) at varying concentrations.
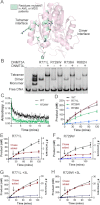
FIGURE 4.
DNMT3A homodimers and homotetramers are active with different mechanisms. Disruption of the dimer interface reduces activity, but the dimers are still active with increased off-rates. The wild type tetramers are labeled gray and the dimer mutants are labeled blue. A and B, _k_cat values of DNMT3A wild type and oligomeric mutants were determined on multisite substrates, poly-dIdC (A), and the human promoter RASSF1A (B.). Below are the wild type (gray) and dimer mutant R882H (blue) time course trace. C, eliminating tetramer formation in DNMT3A increased K _m_DNA (substrate poly-dIdC), above is the bar chart of K _m_DNA values, and below are the kinetic fits. D, DNMT3A homodimers have an increase in off-rate compared with homotetramers, above is the bar chart of _k_off values determined on GCbox30, and below are the kinetic fits. All error bars were determined from at least three experiments given as S.E.; one-way analysis of variance was used to compare wild values to each mutant. *, p > 0.05; **, p > 0.01; ***, p > 0.001.
FIGURE 5.
Dimer interface disrupting mutants eliminate processive catalysis. Chase assays show DNMT3A homotetramers (WT) are processive and dimers (H873A and R882H) are non-processive, A, WT; B, H873A; and C, R882H. ●, only substrate (20 μ
m
bp RASSF1A); ■, substrate and then 400 μ
m
bp chase (pCpGL) at 20 min; ▴, substrate and pCpGL at the start of the reaction. Minimal methylation is detected after addition of chase DNA with the dimer mutant (H873A and R882H), unlike tetramer (WT), which shows less than 10% change in activity. D, homodimers that were formed by disrupting the dimer interface resulted in enzymes that bind DNA followed by methylation and then fast dissociation.
FIGURE 6.
DNMT3L and the formation of heterotetramers restores processivity to R882H DNMT3A. A, the EMSA shows that DNMT3L binds to DNMT3A tetramers to form heterotetramers. R882H binds to DNMT3L; all other mutants at the dimer interface become heterodimers. B, DNMT3L (1:1 ratio) activates DNMT3A tetramers at ∼5-fold. Dimer interface mutants show a 2–4-fold activation other than R882H, which show an 8.8-fold stimulation. DNMT3A heterotetramers (C, WT; D, R882H) are processive, and heterodimers (E, H873A) are non-processive as demonstrated by the processive chase assay. ●, only substrate (20 μ
m
bp RASSF1A); ■, substrate and then 400 μ
m
bp chase (pCpGL at 20 min; ▴, substrate and pCpGL at the start of the reaction.
FIGURE 7.
AML mutants disrupt the tetramer interface and eliminate DNMT3A processivity. A, mutants identified in AML and MDS patients located along DNMT3A tetramer interface. B, AML mutants at the tetramer interface disrupt the homotetramer and form heterotetramer with DNMT3L. C, the rate of catalysis is minimally changed for two of the tetramer interface disrupting mutants (R771L and R729W). D, disruption of the tetramer interface increases the off-rate. E and F, disruption of the tetramer interface also eliminates processivity, as demonstrated by the processive chase assay (E, R771A; F, R729W). G and H, DNMT3L restores processivity by forming a heterotetramer (G, R771A; H, R729W). The chase assay was done as follows: ●, only substrate (20 μ
m
bp RASSF1A); ■, substrate and then 400 μ
m
bp chase (pCpGL) at 20 min; ▴, substrate and pCpGL at the start of the reaction.
Similar articles
- Functional Analysis of DNMT3A DNA Methyltransferase Mutations Reported in Patients with Acute Myeloid Leukemia.
Khrabrova DA, Loiko AG, Tolkacheva AA, Cherepanova NA, Zvereva MI, Kirsanova OV, Gromova ES. Khrabrova DA, et al. Biomolecules. 2019 Dec 18;10(1):8. doi: 10.3390/biom10010008. Biomolecules. 2019. PMID: 31861499 Free PMC article. - DNMT3A in Leukemia.
Brunetti L, Gundry MC, Goodell MA. Brunetti L, et al. Cold Spring Harb Perspect Med. 2017 Feb 1;7(2):a030320. doi: 10.1101/cshperspect.a030320. Cold Spring Harb Perspect Med. 2017. PMID: 28003281 Free PMC article. Review. - AML-Associated Mutations in DNA Methyltransferase DNMT3A.
Khrabrova DA, Yakubovskaya MG, Gromova ES. Khrabrova DA, et al. Biochemistry (Mosc). 2021 Mar;86(3):307-318. doi: 10.1134/S000629792103007X. Biochemistry (Mosc). 2021. PMID: 33838631 Review. - The R882H substitution in the human de novo DNA methyltransferase DNMT3A disrupts allosteric regulation by the tumor supressor p53.
Sandoval JE, Reich NO. Sandoval JE, et al. J Biol Chem. 2019 Nov 29;294(48):18207-18219. doi: 10.1074/jbc.RA119.010827. Epub 2019 Oct 22. J Biol Chem. 2019. PMID: 31640986 Free PMC article. - Mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia patients cause both loss and gain of function and differential regulation by protein partners.
Sandoval JE, Huang YH, Muise A, Goodell MA, Reich NO. Sandoval JE, et al. J Biol Chem. 2019 Mar 29;294(13):4898-4910. doi: 10.1074/jbc.RA118.006795. Epub 2019 Jan 31. J Biol Chem. 2019. PMID: 30705090 Free PMC article.
Cited by
- Base editor scanning charts the DNMT3A activity landscape.
Lue NZ, Garcia EM, Ngan KC, Lee C, Doench JG, Liau BB. Lue NZ, et al. Nat Chem Biol. 2023 Feb;19(2):176-186. doi: 10.1038/s41589-022-01167-4. Epub 2022 Oct 20. Nat Chem Biol. 2023. PMID: 36266353 Free PMC article. - Next Generation Sequencing in MPNs. Lessons from the Past and Prospects for Use as Predictors of Prognosis and Treatment Responses.
Skov V. Skov V. Cancers (Basel). 2020 Aug 6;12(8):2194. doi: 10.3390/cancers12082194. Cancers (Basel). 2020. PMID: 32781570 Free PMC article. Review. - DNMT3A R882 Mutations Confer Unique Clinicopathologic Features in MDS Including a High Risk of AML Transformation.
Jawad M, Afkhami M, Ding Y, Zhang X, Li P, Young K, Xu ML, Cui W, Zhao Y, Halene S, Al-Kali A, Viswanatha D, Chen D, He R, Zheng G. Jawad M, et al. Front Oncol. 2022 Feb 28;12:849376. doi: 10.3389/fonc.2022.849376. eCollection 2022. Front Oncol. 2022. PMID: 35296003 Free PMC article. - The R882H DNMT3A hot spot mutation stabilizes the formation of large DNMT3A oligomers with low DNA methyltransferase activity.
Nguyen TV, Yao S, Wang Y, Rolfe A, Selvaraj A, Darman R, Ke J, Warmuth M, Smith PG, Larsen NA, Yu L, Zhu P, Fekkes P, Vaillancourt FH, Bolduc DM. Nguyen TV, et al. J Biol Chem. 2019 Nov 8;294(45):16966-16977. doi: 10.1074/jbc.RA119.010126. Epub 2019 Oct 3. J Biol Chem. 2019. PMID: 31582562 Free PMC article. - Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns.
Emperle M, Adam S, Kunert S, Dukatz M, Baude A, Plass C, Rathert P, Bashtrykov P, Jeltsch A. Emperle M, et al. Nucleic Acids Res. 2019 Dec 2;47(21):11355-11367. doi: 10.1093/nar/gkz911. Nucleic Acids Res. 2019. PMID: 31620784 Free PMC article.
References
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 - PubMed
- Reik W., Dean W., Walter J. (2001) Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 - PubMed
- Robertson K. D. (2005) DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 - PubMed
- Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 - PubMed
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004) Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical