Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52 - PubMed (original) (raw)
Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52
Tao G Dong et al. Nucleic Acids Res. 2012 Sep.
Abstract
The alternative sigma factor RpoN is an essential colonization factor of Vibrio cholerae and controls important cellular functions including motility and type VI secretion (T6SS). The RpoN regulon has yet to be clearly defined in T6SS-active V. cholerae isolates, which use T6SS to target both bacterial competitors and eukaryotic cells. We hypothesize that T6SS-dependent secreted effectors are co-regulated by RpoN. To systemically identify RpoN-controlled genes, we used chromatin immunoprecipitation coupled with sequencing (ChIP-Seq) and transcriptome analysis (RNA-Seq) to determine RpoN-binding sites and RpoN-controlled gene expression. There were 68 RpoN-binding sites and 82 operons positively controlled by RpoN, among which 37 operons had ChIP-identified binding sites. A consensus RpoN-binding motif was identified with a highly conserved thymine (-14) and an AT-rich region in the middle between the hallmark RpoN-recognized motif GG(-24)/GC(-12). There were seven new RpoN-dependent promoters in the flagellar regions. We identified a small RNA, flaX, downstream of the major flagellin gene flaA. Mutation of flaX substantially reduced motility. In contrast to previous results, we report that RpoN positively regulates the expression of hcp operons and vgrG3 that encode T6SS secreted proteins but has no effect on the expression of the main T6SS cluster encoding sheath and other structural components.
Figures
Figure 1.
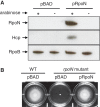
RpoN-3V5 complements the rpoN deletion mutant. The pBAD-derivative plasmid pRpoN carrying arabinose-inducible RpoN-3V5 complements two known RpoN-dependent traits in the rpoN mutant, Hcp expression (A) and motility (B).
Figure 2.
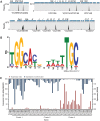
A sample of ChIP-Seq and RNA-Seq data covering the flagellar region and consensus RpoN-binding motif. (A) Genomic structures are shown above the peak panel. The height of each peak corresponds to the number of reads bound to the corresponding ChIP-site. (B) Motif was generated based on all ChIP-peaks using MEME program. The height of each letter represents the occurrence frequency at each location. (C) RNA-Seq data show difference in regulation of flagellar genes by RpoN. The RpoN-dependence (expression ratio) is shown at the bottom with the left axis. The absolute expression for each gene in the rpoN mutant is shown on the top with the right axis. Genes with high-RpoN-dependence and low-expression in the rpoN mutant require RpoN for expression, whereas genes with high-expression in the rpoN mutant likely have other promoters independent of RpoN.
Figure 3.
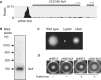
The new sRNA flaX downstream of VC2188. (A) RNA-Seq data show a substantial increase of coverage in the downstream region of flaA. (B) Northern blot analysis confirmed the existence of flaX. (C) Mutation in flaX attenuated motility. (D) Motility of the flaX mutant was complemented by expressing flaX in trans on a sRNA expression vector pNM12. Arabinose (0.02%) was used for inducing the expression of flaX.
Figure 4.
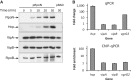
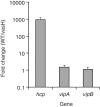
RpoN is required for the expression of Hcp but not VipA or VipB. (A) Protein levels of Hcp, VipA (VCA0107) and VipB (VCA0108) by western blot analysis. The rpoN mutant was transformed with the plasmid pRpoN or the empty vector pBAD18. Cultures were grown aerobically in LB medium at 37°C to exponential phase (OD600 = 0.5), and RpoN was induced by the addition of arabinose (0.1%) for 30 minutes. Samples were taken at different time points and protein expression was detected by western blot analysis. (B) Transcriptional levels of hcp and and vipAB and ChIP-binding of their promoter regions. The rrsA gene encoding 16 S RNA was used as an internal control sample to normalize difference in total RNA quantity among samples for qPCR. VgrG2 is located downstream of hcp2 and used as a control sample for ChIP assay. Monoclonal antibody to RpoN-3V5 was used for ChIP assay.
Figure 5.
VasH, the enhancer-binding protein for RpoN, controls the expression of hcp but not the major cluster genes vipA and vipB. Relative gene expression was compared in wild type V52 and the vasH mutant by qPCR. The 16 S ribosomal RNA gene rrsA was used as a control sample. RNA was extracted in exponential phase cultures OD600 = 0.5.
Similar articles
- Sensing of intracellular Hcp levels controls T6SS expression in Vibrio cholerae.
Manera K, Caro F, Li H, Pei TT, Hersch SJ, Mekalanos JJ, Dong TG. Manera K, et al. Proc Natl Acad Sci U S A. 2021 Jun 22;118(25):e2104813118. doi: 10.1073/pnas.2104813118. Proc Natl Acad Sci U S A. 2021. PMID: 34161288 Free PMC article. - Pleiotropic control of antibiotic biosynthesis, flagellar operon expression, biofilm formation, and carbon source utilization by RpoN in Pseudomonas protegens H78.
Liu Y, Shi H, Wang Z, Huang X, Zhang X. Liu Y, et al. Appl Microbiol Biotechnol. 2018 Nov;102(22):9719-9730. doi: 10.1007/s00253-018-9282-0. Epub 2018 Aug 21. Appl Microbiol Biotechnol. 2018. PMID: 30128583 - Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains.
Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Ishikawa T, et al. PLoS One. 2009 Aug 24;4(8):e6734. doi: 10.1371/journal.pone.0006734. PLoS One. 2009. PMID: 19701456 Free PMC article. - T6SS intraspecific competition orchestrates Vibrio cholerae genotypic diversity.
Kostiuk B, Unterweger D, Provenzano D, Pukatzki S. Kostiuk B, et al. Int Microbiol. 2017 Sep;20(3):130-137. doi: 10.2436/20.1501.01.294. Int Microbiol. 2017. PMID: 29446804 Review. - The Regulatory Functions of σ54 Factor in Phytopathogenic Bacteria.
Yu C, Yang F, Xue D, Wang X, Chen H. Yu C, et al. Int J Mol Sci. 2021 Nov 24;22(23):12692. doi: 10.3390/ijms222312692. Int J Mol Sci. 2021. PMID: 34884502 Free PMC article. Review.
Cited by
- Vibrio cholerae Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System.
Cheng AT, Ottemann KM, Yildiz FH. Cheng AT, et al. PLoS Pathog. 2015 May 22;11(5):e1004933. doi: 10.1371/journal.ppat.1004933. eCollection 2015 May. PLoS Pathog. 2015. PMID: 26000450 Free PMC article. - Microbial herd protection mediated by antagonistic interaction in polymicrobial communities.
Wong M, Liang X, Smart M, Tang L, Moore R, Ingalls B, Dong TG. Wong M, et al. Appl Environ Microbiol. 2016 Dec;82(23):6881-6888. doi: 10.1128/AEM.02210-16. Epub 2016 Sep 16. Appl Environ Microbiol. 2016. PMID: 27637882 Free PMC article. - Defining bacterial regulons using ChIP-seq.
Myers KS, Park DM, Beauchene NA, Kiley PJ. Myers KS, et al. Methods. 2015 Sep 15;86:80-8. doi: 10.1016/j.ymeth.2015.05.022. Epub 2015 May 29. Methods. 2015. PMID: 26032817 Free PMC article. Review. - Double Tubular Contractile Structure of the Type VI Secretion System Displays Striking Flexibility and Elasticity.
Stietz MS, Liang X, Wong M, Hersch S, Dong TG. Stietz MS, et al. J Bacteriol. 2019 Dec 6;202(1):e00425-19. doi: 10.1128/JB.00425-19. Print 2019 Dec 6. J Bacteriol. 2019. PMID: 31636107 Free PMC article. - Microbial Surface Colonization and Biofilm Development in Marine Environments.
Dang H, Lovell CR. Dang H, et al. Microbiol Mol Biol Rev. 2015 Dec 23;80(1):91-138. doi: 10.1128/MMBR.00037-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2015. PMID: 26700108 Free PMC article. Review.
References
- Weil AA, Ivers LC, Harris JB. Cholera: lessons from haiti and beyond. Curr. Infect. Dis. Rep. 2012;14:1–8. - PubMed
- Ritchie JM, Waldor MK. Vibrio cholerae interactions with the gastrointestinal tract: lessons from animal studies. Curr. Top. Microbiol. Immunol. 2009;337:37–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous