De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly - PubMed (original) (raw)
. 2012 Jun 24;44(8):941-5.
doi: 10.1038/ng.2329.
My Huynh, Jennifer L Silhavy, Sangwoo Kim, Tracy Dixon-Salazar, Andrew Heiberg, Eric Scott, Vineet Bafna, Kiley J Hill, Adrienne Collazo, Vincent Funari, Carsten Russ, Stacey B Gabriel, Gary W Mathern, Joseph G Gleeson
Affiliations
- PMID: 22729223
- PMCID: PMC4417942
- DOI: 10.1038/ng.2329
De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly
Jeong Ho Lee et al. Nat Genet. 2012.
Abstract
De novo somatic mutations in focal areas are well documented in diseases such as neoplasia but are rarely reported in malformation of the developing brain. Hemimegalencephaly (HME) is characterized by overgrowth of either one of the two cerebral hemispheres. The molecular etiology of HME remains a mystery. The intractable epilepsy that is associated with HME can be relieved by the surgical treatment hemispherectomy, allowing sampling of diseased tissue. Exome sequencing and mass spectrometry analysis in paired brain-blood samples from individuals with HME (n = 20 cases) identified de novo somatic mutations in 30% of affected individuals in the PIK3CA, AKT3 and MTOR genes. A recurrent PIK3CA c.1633G>A mutation was found in four separate cases. Identified mutations were present in 8-40% of sequenced alleles in various brain regions and were associated with increased neuronal S6 protein phosphorylation in the brains of affected individuals, indicating aberrant activation of mammalian target of rapamycin (mTOR) signaling. Thus HME is probably a genetically mosaic disease caused by gain of function in phosphatidylinositol 3-kinase (PI3K)-AKT3-mTOR signaling.
Figures
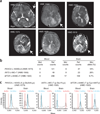
Figure 1
MRI and mutation analysis in hemimegalencephaly. (a) Axial T2-weighted brain MRI of cases identified with mutations. Arrows indicate the affected hemispheres, showing thickened cortical mantle, changes in white matter signal and alterations in ventricular shape, resulting in increased hemispheric size and midline shift of falx cerebri. (b) Sequencing counts from exome sequencing of each of three brain-blood paired samples. Mut, mutation; ref, reference. (c) Mass spectrometry validation of mutations. Wild-type sequences (blue) and de novo mutations (red) are correlated with results from next-generation sequencing.
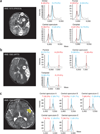
Figure 2
Consistent de novo mutation burden across affected hemispheres. (a) HME-1573 sampled during surgery in left orbital (Orb), frontal (Fr), central operculum (Co) and occipital (Occ) regions. (b) HME-1656 sampled in parietal (Par), central operculum and temporal (Tem) regions. (c) HME-1563 was sampled multiple independent times in the central operculum region (yellow circles), but samples could not be precisely aligned anatomically. Left, MRI scans; circles indicate brain areas examined by mass spectrometry analysis. Right, each sample aliquot was analyzed by mass spectrometry for mutation burden, describing variation in mutation burden across anatomical locations.
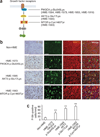
Figure 3
The de novo mutations identified in HME correlate with hyperactive mTOR signaling. (a) Schematic of the PI3K-AKT3-mTOR pathway and downstream phosphorylated ribosomal protein S6 kinase (P-S6K) and phosphorylated ribosomal protein S6 (P-S6). (b) HME pathological samples show an increased percentage of cells with positive staining for P-S6 and MAP2. Left P-S6, biotin-streptavidin DAB staining; right, P-S6, fluorescence-conjugated staining. MAP2 was used to identify neurons. Scale bars, 100 µm. (c) Quantification of P-S6–positive cells from 5–7 representative cortical areas per case. **P < 0.01 (relative to non-HME samples, one-way ANOVA with Bonferroni posttest, _n_ > 50 cells per region). Error bars, s.e.m.
Similar articles
- Profiling PI3K-AKT-MTOR variants in focal brain malformations reveals new insights for diagnostic care.
Pirozzi F, Berkseth M, Shear R, Gonzalez L, Timms AE, Sulc J, Pao E, Oyama N, Forzano F, Conti V, Guerrini R, Doherty ES, Saitta SC, Lockwood CM, Pritchard CC, Dobyns WB, Novotny E, Wright JNN, Saneto RP, Friedman S, Hauptman J, Ojemann J, Kapur RP, Mirzaa GM. Pirozzi F, et al. Brain. 2022 Apr 29;145(3):925-938. doi: 10.1093/brain/awab376. Brain. 2022. PMID: 35355055 Free PMC article. - PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia.
Jansen LA, Mirzaa GM, Ishak GE, O'Roak BJ, Hiatt JB, Roden WH, Gunter SA, Christian SL, Collins S, Adams C, Rivière JB, St-Onge J, Ojemann JG, Shendure J, Hevner RF, Dobyns WB. Jansen LA, et al. Brain. 2015 Jun;138(Pt 6):1613-28. doi: 10.1093/brain/awv045. Epub 2015 Feb 25. Brain. 2015. PMID: 25722288 Free PMC article. - Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia.
D'Gama AM, Geng Y, Couto JA, Martin B, Boyle EA, LaCoursiere CM, Hossain A, Hatem NE, Barry BJ, Kwiatkowski DJ, Vinters HV, Barkovich AJ, Shendure J, Mathern GW, Walsh CA, Poduri A. D'Gama AM, et al. Ann Neurol. 2015 Apr;77(4):720-5. doi: 10.1002/ana.24357. Epub 2015 Feb 26. Ann Neurol. 2015. PMID: 25599672 Free PMC article. - Hemimegalencephaly, a paradigm for somatic postzygotic neurodevelopmental disorders.
Baek ST, Gibbs EM, Gleeson JG, Mathern GW. Baek ST, et al. Curr Opin Neurol. 2013 Apr;26(2):122-7. doi: 10.1097/WCO.0b013e32835ef373. Curr Opin Neurol. 2013. PMID: 23449172 Review. - mTOR signaling in epilepsy: insights from malformations of cortical development.
Crino PB. Crino PB. Cold Spring Harb Perspect Med. 2015 Apr 1;5(4):a022442. doi: 10.1101/cshperspect.a022442. Cold Spring Harb Perspect Med. 2015. PMID: 25833943 Free PMC article. Review.
Cited by
- When, where and which PIK3CA mutations are pathogenic in congenital disorders.
Angulo-Urarte A, Graupera M. Angulo-Urarte A, et al. Nat Cardiovasc Res. 2022 Aug;1(8):700-714. doi: 10.1038/s44161-022-00107-8. Epub 2022 Aug 8. Nat Cardiovasc Res. 2022. PMID: 39196083 Review. - The molecular genetics of PI3K/PTEN/AKT/mTOR pathway in the malformations of cortical development.
Ma Q, Chen G, Li Y, Guo Z, Zhang X. Ma Q, et al. Genes Dis. 2023 Jul 16;11(5):101021. doi: 10.1016/j.gendis.2023.04.041. eCollection 2024 Sep. Genes Dis. 2023. PMID: 39006182 Free PMC article. Review. - Mapping recurrent mosaic copy number variation in human neurons.
Sun C, Kathuria K, Emery SB, Kim B, Burbulis IE, Shin JH; Brain Somatic Mosaicism Network; Weinberger DR, Moran JV, Kidd JM, Mills RE, McConnell MJ. Sun C, et al. Nat Commun. 2024 May 17;15(1):4220. doi: 10.1038/s41467-024-48392-0. Nat Commun. 2024. PMID: 38760338 Free PMC article. - CNS autoimmune response in the MAM/pilocarpine rat model of epileptogenic cortical malformation.
Costanza M, Ciotti A, Consonni A, Cipelletti B, Cattalini A, Cagnoli C, Baggi F, de Curtis M, Colciaghi F. Costanza M, et al. Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2319607121. doi: 10.1073/pnas.2319607121. Epub 2024 Apr 18. Proc Natl Acad Sci U S A. 2024. PMID: 38635635 Free PMC article. - Malformations of cortical development: Fetal imaging and genetics.
Wang LL, Pan PS, Ma H, He C, Qin ZL, He W, Huang J, Tan SY, Meng DH, Wei HW, Yin AH. Wang LL, et al. Mol Genet Genomic Med. 2024 Apr;12(4):e2440. doi: 10.1002/mgg3.2440. Mol Genet Genomic Med. 2024. PMID: 38634212 Free PMC article.
References
- Salamon N, et al. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain. 2006;129:352–365. - PubMed
- Di Rocco C, Battaglia D, Pietrini D, Piastra M, Massimi L. Hemimegalencephaly: clinical implications and surgical treatment. Childs Nerv. Syst. 2006;22:852–866. - PubMed
- Crino PB. mTOR: a pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011;17:734–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 NS047101/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 NS048453/NS/NINDS NIH HHS/United States
- R01 NS038992/NS/NINDS NIH HHS/United States
- R01 NS083823/NS/NINDS NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P30NS047101/NS/NINDS NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- R01 NS041537/NS/NINDS NIH HHS/United States
- R01 NS052455/NS/NINDS NIH HHS/United States
- P01 HD070494/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous