Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae - PubMed (original) (raw)
. 2012 Sep 1;40(17):8646-61.
doi: 10.1093/nar/gks609. Epub 2012 Jun 26.
Affiliations
- PMID: 22735702
- PMCID: PMC3458554
- DOI: 10.1093/nar/gks609
Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae
Jason Talkish et al. Nucleic Acids Res. 2012.
Abstract
To better define the roles of assembly factors required for eukaryotic ribosome biogenesis, we have focused on one specific step in maturation of yeast 60 S ribosomal subunits: processing of 27SB pre-ribosomal RNA. At least 14 assembly factors, the 'B-factor' proteins, are required for this step. These include most of the major functional classes of assembly factors: RNA-binding proteins, scaffolding protein, DEAD-box ATPases and GTPases. We have investigated the mechanisms by which these factors associate with assembling ribosomes. Our data establish a recruitment model in which assembly of the B-factors into nascent ribosomes ultimately leads to the recruitment of the GTPase Nog2. A more detailed analysis suggests that this occurs in a hierarchical manner via two largely independent recruiting pathways that converge on Nog2. Understanding recruitment has allowed us to better determine the order of association of all assembly factors functioning in one step of ribosome assembly. Furthermore, we have identified a novel subcomplex composed of the B-factors Nop2 and Nip7. Finally, we identified a means by which this step in ribosome biogenesis is regulated in concert with cell growth via the TOR protein kinase pathway. Inhibition of TOR kinase decreases association of Rpf2, Spb4, Nog1 and Nog2 with pre-ribosomes.
Figures
Figure 1.
Pathway for processing of pre-rRNA and maturation of pre-rRNPs in Saccharomyces cerevisiae. (A) rRNA processing pathway. The 35S primary transcript, synthesized by RNA polymerase I, contains sequences for mature 18S, 25S and 5.8S rRNA flanked and separated by external and internal transcribed spacers (ETS/ITS). The 5S rRNA is transcribed from separate linked genes by RNA polymerase III and contains a 3′ETS. The spacers are removed by a series of endonucleolytic and exonucleolytic processing steps in assembling ‘_pre-rRNPs_’. Processing of the 27SB pre-rRNA to form 25.5S and 7S pre-rRNAs involves endonucleolytic cleavage at the C2 site in ITS2, as shown (boxed). (B) Assembly intermediates in the biogenesis of 60S ribosomal subunits. ‘pre-rRNPs’ that mature into 60S ribosomal subunits are shown, with the pre-rRNAs contained within them indicated. Known numbers of assembly factors required for each of the consecutive steps in subunit maturation are shown.
Figure 2.
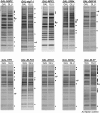
Depletion of B-factors causes few changes in the SDS–PAGE profile of proteins contained in pre-ribosomes. Nine of the 12 B-factors necessary for processing of 27SB pre-rRNA were individually depleted using the conditional GAL promoter. TAP-tagged ribosome assembly factor Nop7 was used to purify 90S and 66S pre-ribosomes from yeast grown in YEPGal (left lane in each pair), and from strains grown in YEPGal and shifted to YEPGlu for 16 h to deplete each B-factor (right lane in each pair). Proteins present in purified pre-ribosomes were separated by SDS–PAGE and stained with silver. TAP-tagged Rpf2 was used to purify pre-ribosomes from the GAL-RLP7 strain in which Rlp7 was depleted. Rlp7 is required for processing of 27SA3 pre-rRNA. Thus, this strain serves as a control to demonstrate specificity of the gel profiles for B-factor-depleted strains. Silver-stained bands that increased in the absence of each assembly factor are indicated with solid circles. Bands that decreased are indicated with hollow circles. When able to be identified, the protein under control of the GAL promoter is indicated with #.
Figure 3.
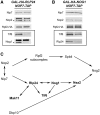
All the B-factors are necessary to recruit Nog2 to pre-ribosomes; only a subset of B-factors are required to recruit Nsa2 into pre-ribosomes. (A) The changes in the levels of pre-ribosome-associated Nsa2 and Nog2 in the absence of each of the B-factors. Pre-ribosomes were purified from cells grown in YEPGal or shifted to YEPGlu to deplete each B-factor. Proteins from pre-rRNPs were resolved by SDS–PAGE and assayed by western blotting. In each pair of samples shown in this figure and in Figures 4–7, protein from the undepleted strain is on the left and protein from the depleted strain is on the right. In this blot and all subsequent blots, where two bands are detected, the bona fide protein is indicated with an asterisk. Nop7-TAP serves as the loading control. (B) Nog2 is not required to recruit any of the B-factors. Proteins in pre-ribosomes purified from GAL-NOG2 yeast grown in YEPGal, or shifted to YEPGlu to deplete Nog2 were assayed by western blotting. Nop7-TAP serves as a loading control. Each pair of samples is shown as described in Figure 3A. (C) All of the B-factors are required to recruit Nog2 to pre-ribosomes. A subset of B-factors is required to recruit Nsa2 and Nog2.
Figure 4.
Spb4 depends on Rpf2 to dock with pre-ribosomes. (A) Rpf2 is necessary to load Spb4 into nascent ribosomes. Pre-ribosomes were purified from a GAL-RPF2 strain grown in YEPGal or shifted to YEPGlu to deplete Rpf2. Pre-ribosomes were purified from whole cell lysates of undepleted and depleted cells using TAP-tagged Nop7. Proteins from pre-rRNPs were resolved by SDS–PAGE and assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Rpf2 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. B-factors that decrease in the absence of Rpf2 are boxed. (B) No other B-factors are recruited into pre-ribosomes by Spb4. Proteins in pre-ribosomes purified from GAL-SPB4 yeast grown in YEPGal or shifted to YEPGlu to deplete Spb4 were assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Spb4 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (C) Hierarchy for recruiting Rpf2, Spb4 and Nog2 into pre-ribosomes.
Figure 5.
Nop2 and Nip7 form a subcomplex, are interdependent for assembly into pre-ribosomes and function in recruiting of other B-factors. (A) Nop2 and Nip7 form a heteromer in vivo. Yeast strains expressing TAP-tagged Nop2 or Nip7 were grown in YEPGlu medium to 3 × 107 cells/ml. Whole-cell extracts from the NOP2-TAP strain (lane 1) or the supernatants after high-speed centrifugation (lane 2) were subjected to affinity purification using Nop2-TAP, to purify Nop2-containing pre-ribosomes or subcomplexes, respectively. Purified proteins were identified by mass spectrometry. Proteins labeled with ‘#’ are common contaminants in purified samples. Whole-cell extract from the NIP7-TAP strain was subjected to affinity purification using Nip7-TAP (lane 3). Nop2 and Nip7 in these purified samples were identified by mass spectrometry. (B and C) Nop2 and Nip7 are interdependent for their assembly into pre-rRNPs and important for incorporation of other B-factors into pre-ribosomes. GAL-nip7-1 or GAL-NOP2 yeast were grown in YEPGal or shifted to YEPGlu. Whole-cell extracts were subjected to affinity purification, using Nop7-TAP to isolate pre-ribosomes. Amounts of B-factors present in the purified pre-ribosomes were analyzed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Nop2 or Nip7 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (D) Nop2 and Nip7 are important for the stable association of all the B-factors.
Figure 6.
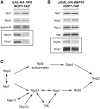
Nog1 and Rlp24 are important for association with pre-ribosomes of Tif6, as well as each other. (A and B) The changes in amounts of pre-ribosomal proteins when either Rlp24 or Nog1 is depleted. GAL-RLP24 or GAL-NOG1 yeast were grown in YEPGal or shifted to YEPGlu for 16 h to deplete either assembly factor. Pre-ribosomes were purified from whole-cell lysates of undepleted and depleted cells using TAP-tagged Nop7. Western blotting was used to assay amounts of pre-ribosomal proteins. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Nog1 or Rlp24 from preribosomes, and the Nop7-TAP loading control are shown in Figure 3A. (C) Interdependence among Rlp24, Nog1, Tif6, Nsa2 and Nog2 for assembly into pre-ribosomes. Proteins in boldface were previously studied by Fromont-Racine and coworkers.
Figure 7.
Both Tif6 and Dbp10 are required for assembly of Rlp24 and Nog1 into pre-ribosomes.(A) Tif6 is required to recruit Rlp24 and Nog1 pre-rRNPs. Proteins present in pre-rRNPs purified from a GAL-TIF6 strain grown in YEPGal or shifted to YEPGlu for 16 h were resolved by SDS–PAGE and subjected to western blot analysis. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Tif6 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (B) Dbp10 is important for association of Rlp24 and Nog1 with pre-rRNPs. The GAL-DBP10 strain was grown in YEPGal or shifted to YEPGlu for 16 h to deplete Dbp10. Pre-ribosomes were purified from extracts prepared from each strain, and proteins were resolved by SDS–PAGE. Pre-ribosomal proteins were assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Dbp10 from preribosomes, and the Nop7-TAP loading control are shown in Figure 3A. (C) Position in the association hierarchy for Tif6 and Dbp10.
Figure 8.
The TOR protein kinase pathway regulates assembly of 60S ribosomal subunits via targeting the B-factors Rpf2, Spb4, Nog1 and Nog2. The Nop7-TAP strain was grown at 30°C in YEPGlu medium to 3 × 107 cells/ml. Two more cultures were grown to 2 × 107 cells/ml, and 200 ng/ml rapamycin was added to each culture. These two cultures were then grown for another 1 or 2.5 h to 3 × 107 cells/ml. Whole-cell extracts were subjected to single-step purification using TAP-tagged Nop7 to isolate 66S pre-ribosomes. Proteins present in the affinity-purified pre-ribosomes were resolved by SDS–PAGE and stained with silver (lanes 1–3) or subjected to western blot analysis (lanes 4–6). Proteins from untreated cells are in lanes 1 and 4, those from cells treated with rapamycin for 1 h are in lanes 2 and 5 and those from cells treated for 2.5 h are in lanes 3 and 6. Nop7-TAP serves as the loading control. Proteins with decreased association with pre-ribosomes upon treatment with rapamycin are boxed. The pre-rRNA processing step in which each protein tested functions is indicated.
Figure 9.
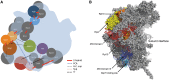
Physical and genetic interactions provide clues about the location of the B-factors in pre-ribosomes. (A) Predicted locations of B-factors in assembling 60S subunits. Binding sites of proteins were predicted using a combination of genetic and physical interactions and then superimposed on a schematic of the 60S subunit viewed form the intersubunit side. Grey circles indicate assembly factors and ribosomal proteins that were used as landmarks to help position the B-factors. Genetic interactions listed in
Supplementary Table S3
are indicated. PCA, protein complementation assay; H.C. sup, high-copy suppressor; Sup, extragenic suppressor; sl−, synthetic lethal. (B) r-proteins such as L4, L15 and L18 are shown in the crystal structure of the 60S subunit as viewed from the crown view of the intersubunit face rotated right 90°. Domains I and III of 25S rRNA are indicated in red and blue, respectively. The Nop7 binding site determined by Granneman et al. (56) is shown in cyan. Pymol images of the ribosome structure were generated using PDB files 3U5H and 3U5I.
Similar articles
- Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing.
Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL Jr. Gamalinda M, et al. Nucleic Acids Res. 2013 Feb 1;41(3):1965-83. doi: 10.1093/nar/gks1272. Epub 2012 Dec 24. Nucleic Acids Res. 2013. PMID: 23268442 Free PMC article. - Ribosomal proteins L7 and L8 function in concert with six A₃ assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits.
Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL Jr. Jakovljevic J, et al. RNA. 2012 Oct;18(10):1805-22. doi: 10.1261/rna.032540.112. Epub 2012 Aug 14. RNA. 2012. PMID: 22893726 Free PMC article. - Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits.
Dembowski JA, Kuo B, Woolford JL Jr. Dembowski JA, et al. Nucleic Acids Res. 2013 Sep;41(16):7889-904. doi: 10.1093/nar/gkt545. Epub 2013 Jun 20. Nucleic Acids Res. 2013. PMID: 23788678 Free PMC article. - Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast.
Konikkat S, Woolford JL Jr. Konikkat S, et al. Biochem J. 2017 Jan 15;474(2):195-214. doi: 10.1042/BCJ20160516. Biochem J. 2017. PMID: 28062837 Free PMC article. Review. - Ribosome biogenesis in the yeast Saccharomyces cerevisiae.
Woolford JL Jr, Baserga SJ. Woolford JL Jr, et al. Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review.
Cited by
- Viewing pre-60S maturation at a minute's timescale.
Zisser G, Ohmayer U, Mauerhofer C, Mitterer V, Klein I, Rechberger GN, Wolinski H, Prattes M, Pertschy B, Milkereit P, Bergler H. Zisser G, et al. Nucleic Acids Res. 2018 Apr 6;46(6):3140-3151. doi: 10.1093/nar/gkx1293. Nucleic Acids Res. 2018. PMID: 29294095 Free PMC article. - Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes.
Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, Ma C, Lei J, Dong MQ, Woolford JL Jr, Gao N. Wu S, et al. Nature. 2016 Jun 2;534(7605):133-7. doi: 10.1038/nature17942. Epub 2016 May 25. Nature. 2016. PMID: 27251291 Free PMC article. - Pol5 is an essential ribosome biogenesis factor required for 60S ribosomal subunit maturation in Saccharomyces cerevisiae.
Ramos-Sáenz A, González-Álvarez D, Rodríguez-Galán O, Rodríguez-Gil A, Gaspar SG, Villalobo E, Dosil M, de la Cruz J. Ramos-Sáenz A, et al. RNA. 2019 Nov;25(11):1561-1575. doi: 10.1261/rna.072116.119. Epub 2019 Aug 14. RNA. 2019. PMID: 31413149 Free PMC article. - RNA folding and functions of RNA helicases in ribosome biogenesis.
Mitterer V, Pertschy B. Mitterer V, et al. RNA Biol. 2022 Jan;19(1):781-810. doi: 10.1080/15476286.2022.2079890. RNA Biol. 2022. PMID: 35678541 Free PMC article. Review.
References
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. - PubMed
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases