A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity - PubMed (original) (raw)
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity
Martin Jinek et al. Science. 2012.
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using CRISPR RNAs (crRNAs) to guide the silencing of invading nucleic acids. We show here that in a subset of these systems, the mature crRNA that is base-paired to trans-activating crRNA (tracrRNA) forms a two-RNA structure that directs the CRISPR-associated protein Cas9 to introduce double-stranded (ds) breaks in target DNA. At sites complementary to the crRNA-guide sequence, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the noncomplementary strand. The dual-tracrRNA:crRNA, when engineered as a single RNA chimera, also directs sequence-specific Cas9 dsDNA cleavage. Our study reveals a family of endonucleases that use dual-RNAs for site-specific DNA cleavage and highlights the potential to exploit the system for RNA-programmable genome editing.
Figures
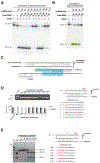
Fig. 1.. Cas9 is a DNA endonuclease guided by two RNA molecules.
(A) Cas9 was programmed with a 42-nucleotide crRNA-sp2 (crRNA containing spacer 2 sequence) in the presence or absence of 75-nucleotide tracrRNA. The complex was added to circular or XhoI-linearized plasmid DNA bearing a sequence complementary to spacer 2 and a functional PAM. crRNA-sp1, specificity control; M, DNA marker; refer to fig. S3A. (B) Cas9 was programmed with crRNA-sp2 and tracrRNA (nucleotides 4-89). The complex was incubated with double- or single-stranded DNAs harboring a sequence complementary to spacer 2 and a functional PAM (4). The complementary or non-complementary strands of the DNA were 5’-radiolabeled and annealed with a non-labeled partner strand. Refer to fig. S3B, C. (C) Sequencing analysis of cleavage products from Fig. 1A. Termination of primer extension in the sequencing reaction indicates the position of the cleavage site. The 3’ terminal A overhang (asterisk) is an artifact of the sequencing reaction. Refer to fig. S5A, C. (D) The cleavage products from Fig. 1B were analyzed alongside 5’ end-labeled size markers derived from the complementary and non-complementary strands of the target DNA duplex. M, marker; P, cleavage product. Refer to fig. S5B, C. (E) Schematic representation of tracrRNA, crRNA-sp2 and protospacer 2 DNA sequences; regions of crRNA complementarity to tracrRNA (orange) and the protospacer DNA (yellow) are represented; PAM sequence, grey; cleavage sites mapped in (C) and (D) are represented by blue arrows (C), a red arrow (D, complementary strand) and a red line (D, non-complementary strand).
Fig. 2.. Cas9 uses two nuclease domains to cleave the two strands in the target DNA.
(A) Top: Schematic representation of Cas9 domain structure showing the positions of domain mutations. Bottom: Complexes of wild-type or nuclease mutant Cas9 proteins with tracrRNA:crRNA-sp2 were assayed for endonuclease activity as in Fig. 1A. (B) Complexes of wild-type Cas9 or nuclease domain mutants with tracrRNA and crRNA-sp2 were tested for activity as in Fig. 1B.
Fig. 3.. Cas9-catalyzed cleavage of target DNA requires an activating domain in tracrRNA and is governed by a seed sequence in the crRNA.
(A) Cas9-tracrRNA:crRNA complexes were reconstituted using 42-nucleotide crRNA-sp2 and truncated tracrRNA constructs and assayed for cleavage activity as in Fig. 1B. (B) Cas9 programmed with full-length tracrRNA and crRNA-sp2 truncations was assayed for activity as in (A). (C) Minimal regions of tracrRNA and crRNA capable of guiding Cas9-mediated DNA cleavage (blue box). (D) Plasmids containing wild-type or mutant protospacer 2 sequences with indicated point mutations (right) were cleaved in vitro by programmed Cas9 as in Fig. 1A (left top) and used for transformation assays of wild-type or pre-crRNA-deficient S. pyogenes (left bottom). The transformation efficiency was calculated as CFU per μg of plasmid DNA; error bars represent standard deviations for three biological replicates. (E) Plasmids containing wild-type and mutant protospacer 2 inserts with varying extent of crRNA-target DNA mismatches (right) were cleaved in vitro by programmed Cas9 (left). The cleavage reactions were further digested with XmnI. The 1880 bp and 800 bp fragments are Cas9-generated cleavage products.
Fig. 4.. A PAM is required to license target DNA cleavage by the Cas9-tracrRNA:crRNA complex.
(A) Dual RNA-programmed Cas9 was tested for activity as in Fig. 1B. Wild-type and mutant PAM sequences in target DNAs are indicated (right). (B) Protospacer 4 target DNA duplexes (labeled at both 5’ ends) containing wild-type and mutant PAM motifs were incubated with Cas9 programmed with tracrRNA (nt 23-89):crRNA-sp4. At indicated time points (min), aliquots of the cleavage reaction were taken and analyzed as in Fig. 1B. (C) Electrophoretic mobility shift assays were performed using RNA-programmed Cas9 (D10A/H840A) and protospacer 4 target DNA duplexes (same as in (B)) containing wild-type and mutated PAM motifs. Cas9 (D10A/H840A)-RNA complex was titrated from 100 pM to 1 μM.
Fig. 5.. Cas9 can be programmed using a single engineered RNA molecule combining tracrRNA and crRNA features.
(A) Top: In Type II CRISPR/Cas systems, Cas9 is guided by a two-RNA structure formed by activating tracrRNA and targeting crRNA to cleave site-specifically target dsDNA (refer to fig. S1). Bottom: A chimeric RNA generated by fusing the 3’ end of crRNA to the 5’ end of tracrRNA. (B) A plasmid harboring protospacer 4 target sequence and a wild-type PAM was subjected to cleavage by Cas9 programmed with tracrRNA(4-89):crRNA-sp4 duplex or _in vitro_-transcribed chimeric RNAs constructed by joining the 3’ end of crRNA to the 5’ end of tracrRNA with a GAAA tetraloop. Cleavage reactions were analyzed by restriction mapping with XmnI. Sequences of chimeric RNAs A and B are shown with DNA-targeting (yellow), crRNA repeat-derived (orange) and tracrRNA-derived (light blue) sequences. (C) Protospacer 4 DNA duplex cleavage reactions were performed as in Fig. 1B. (D) Five chimeric RNAs designed to target the GFP gene were used to program Cas9 to cleave a GFP gene-containing plasmid. Plasmid cleavage reactions were performed as in Fig. 3E, except that the plasmid DNA was restriction mapped with AvrII following Cas9 cleavage.
Comment in
- Molecular biology. A Swiss army knife of immunity.
Brouns SJ. Brouns SJ. Science. 2012 Aug 17;337(6096):808-9. doi: 10.1126/science.1227253. Science. 2012. PMID: 22904002 No abstract available. - RNA-mediated programmable DNA cleavage.
Barrangou R. Barrangou R. Nat Biotechnol. 2012 Sep;30(9):836-8. doi: 10.1038/nbt.2357. Nat Biotechnol. 2012. PMID: 22965054 No abstract available. - Pioneers of revolutionary CRISPR gene editing win chemistry Nobel.
Ledford H, Callaway E. Ledford H, et al. Nature. 2020 Oct;586(7829):346-347. doi: 10.1038/d41586-020-02765-9. Nature. 2020. PMID: 33028993 No abstract available.
Similar articles
- The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. Fonfara I, et al. Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362 - Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria.
Gasiunas G, Barrangou R, Horvath P, Siksnys V. Gasiunas G, et al. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. doi: 10.1073/pnas.1208507109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949671 Free PMC article. - The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems.
Chylinski K, Le Rhun A, Charpentier E. Chylinski K, et al. RNA Biol. 2013 May;10(5):726-37. doi: 10.4161/rna.24321. Epub 2013 Apr 5. RNA Biol. 2013. PMID: 23563642 Free PMC article. - CRISPR-Cas9 Structures and Mechanisms.
Jiang F, Doudna JA. Jiang F, et al. Annu Rev Biophys. 2017 May 22;46:505-529. doi: 10.1146/annurev-biophys-062215-010822. Epub 2017 Mar 30. Annu Rev Biophys. 2017. PMID: 28375731 Review. - Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.
Charpentier E, Richter H, van der Oost J, White MF. Charpentier E, et al. FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review.
Cited by
- Development of an optimized protocol for generating knockout cancer cell lines using the CRISPR/Cas9 system, with emphasis on transient transfection.
Mousavi Kahaki SA, Ebrahimzadeh N, Fahimi H, Moshiri A. Mousavi Kahaki SA, et al. PLoS One. 2024 Nov 14;19(11):e0310368. doi: 10.1371/journal.pone.0310368. eCollection 2024. PLoS One. 2024. PMID: 39541329 Free PMC article. - End-point diagnostics of Giardia duodenalis assemblages A and B by combining RPA with CRISPR/Cas12a from human fecal samples.
Wang Y, Yu F, Fu Y, Zhang Q, Zhao J, Qin Z, Shi K, Wu Y, Li J, Li X, Zhang L. Wang Y, et al. Parasit Vectors. 2024 Nov 12;17(1):463. doi: 10.1186/s13071-024-06559-0. Parasit Vectors. 2024. PMID: 39533301 Free PMC article. - Engineering structural variants to interrogate genome function.
Koeppel J, Weller J, Vanderstichele T, Parts L. Koeppel J, et al. Nat Genet. 2024 Nov 12. doi: 10.1038/s41588-024-01981-7. Online ahead of print. Nat Genet. 2024. PMID: 39533047 Review. - EvoAI enables extreme compression and reconstruction of the protein sequence space.
Ma Z, Li W, Shen Y, Xu Y, Liu G, Chang J, Li Z, Qin H, Tian B, Gong H, Liu DR, Thuronyi BW, Voigt CA, Zhang S. Ma Z, et al. Nat Methods. 2024 Nov 11. doi: 10.1038/s41592-024-02504-2. Online ahead of print. Nat Methods. 2024. PMID: 39528677 - Proliferation Analyses of Conditional Knockdown Strains Using CRISPR Interference in Fission Yeast.
Ishikawa K, Saitoh S. Ishikawa K, et al. Methods Mol Biol. 2025;2862:171-186. doi: 10.1007/978-1-0716-4168-2_12. Methods Mol Biol. 2025. PMID: 39527200
References
- Wiedenheft B, Sternberg SH, Doudna JA, Nature 482, 331–338 (2012). - PubMed
- Bhaya D, Davison M, Barrangou R, Annu. Rev. Genet. 45, 273–297 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials