Estimating kinship in admixed populations - PubMed (original) (raw)
Estimating kinship in admixed populations
Timothy Thornton et al. Am J Hum Genet. 2012.
Abstract
Genome-wide association studies (GWASs) are commonly used for the mapping of genetic loci that influence complex traits. A problem that is often encountered in both population-based and family-based GWASs is that of identifying cryptic relatedness and population stratification because it is well known that failure to appropriately account for both pedigree and population structure can lead to spurious association. A number of methods have been proposed for identifying relatives in samples from homogeneous populations. A strong assumption of population homogeneity, however, is often untenable, and many GWASs include samples from structured populations. Here, we consider the problem of estimating relatedness in structured populations with admixed ancestry. We propose a method, REAP (relatedness estimation in admixed populations), for robust estimation of identity by descent (IBD)-sharing probabilities and kinship coefficients in admixed populations. REAP appropriately accounts for population structure and ancestry-related assortative mating by using individual-specific allele frequencies at SNPs that are calculated on the basis of ancestry derived from whole-genome analysis. In simulation studies with related individuals and admixture from highly divergent populations, we demonstrate that REAP gives accurate IBD-sharing probabilities and kinship coefficients. We apply REAP to the Mexican Americans in Los Angeles, California (MXL) population sample of release 3 of phase III of the International Haplotype Map Project; in this sample, we identify third- and fourth-degree relatives who have not previously been reported. We also apply REAP to the African American and Hispanic samples from the Women's Health Initiative SNP Health Association Resource (WHI-SHARe) study, in which hundreds of pairs of cryptically related individuals have been identified.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Kinship Coefficients Plotted against Zero-IBD-Sharing Probabilities Estimated kinship coefficients plotted against zero-IBD-sharing-probability estimates for three population-structure settings. (A, C, and E) Scatter plots comparing the REAP kinship-coefficient estimator from Equation 3 with the REAP zero-IBD-sharing-probability estimator from Equation 4 for population-structure settings 1 (A), 2 (C), and 3 (E). (B, D, and F) Scatter plots comparing the homogeneous-population kinship-coefficient estimator from Equation 1 with the homogeneous-population zero-IBD-sharing-probability estimator from Equation 2 for population-structure settings 1 (B), 2 (D), and 3 (F). Zero-IBD-sharing-probability and kinship-coefficient estimates were calculated with 10,000 simulated random SNPs.
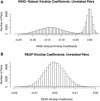
Figure 2
KING-Robust and REAP Kinship-Coefficient Histograms for Unrelated Pairs with Admixture (A and B) Histograms of kinship coefficients estimated with the KING-robust kinship-coefficient estimator (A) and the REAP kinship-coefficient estimator from Equation 3 (B) for all pairs of unrelated individuals in population-structure setting 2. The vertical line at 0 in each histogram represents the true kinship coefficient for all pairs. Kinship-coefficient estimates were calculated with 10,000 simulated random SNPs.
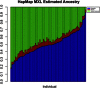
Figure 3
Individual-Ancestry Estimates for HapMap MXL Individual-ancestry estimates for 86 HapMap MXL sample individuals from a supervised structure analysis with the frappe software program. In the figure, each individual is represented by a vertical bar; European (HapMap CEU) and African (HapMap YRI) ancestry contributions are in blue and red, respectively, and Native American (HGDP samples from the Americas) ancestry contributions are in green.
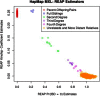
Figure 4
REAP Kinship Coefficients versus Zero-IBD-Sharing Probabilities for HapMap MXL REAP kinship-coefficient estimates are plotted against REAP zero-IBD-sharing-probability estimates for the HapMap MXL sample. REAP estimates were calculated with the kinship-coefficient and zero-IBD-sharing-probability estimators from Equations 3 and 4, respectively. Relative pairs were classified on the basis of kinship-coefficient and zero-IBD-sharing-probability estimates.
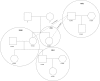
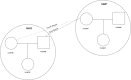
Figure 5
Example of an Extended Pedigree Reconstructed with REAP in HapMap MXL REAP-inferred pedigree relationships for four HapMap-reported pedigrees from the MXL sample are given. HapMap-reported pedigree relationships are circled, and HapMap-reported pedigree identification numbers (M008, 2382, M011, and M012) are given in bold font in each of the circles.
Figure 6
Example of Two HapMap MXL Pedigrees Connected with REAP Pedigree relationships for two HapMap-reported pedigrees from the MXL sample are given. HapMap-reported pedigree relationships are circled, and HapMap-reported pedigree identification numbers (M007 and M032) are given in bold font in each of the circles.
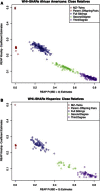
Figure 7
REAP Kinship Coefficients versus Zero-IBD-Sharing Probabilities for WHI-SHARe (A and B) REAP kinship-coefficient estimates are plotted against REAP zero-IBD-sharing-probability estimates for the WHI-SHARe self-reported African Americans and self-reported Hispanics, respectively. REAP estimates were calculated with the kinship-coefficient and zero-IBD-sharing-probability estimators from Equations 3 and 4, respectively.
Similar articles
- Detecting Heterogeneity in Population Structure Across the Genome in Admixed Populations.
McHugh C, Brown L, Thornton TA. McHugh C, et al. Genetics. 2016 Sep;204(1):43-56. doi: 10.1534/genetics.115.184184. Epub 2016 Jul 20. Genetics. 2016. PMID: 27440868 Free PMC article. - Genome-wide Significance Thresholds for Admixture Mapping Studies.
Grinde KE, Brown LA, Reiner AP, Thornton TA, Browning SR. Grinde KE, et al. Am J Hum Genet. 2019 Mar 7;104(3):454-465. doi: 10.1016/j.ajhg.2019.01.008. Epub 2019 Feb 14. Am J Hum Genet. 2019. PMID: 30773276 Free PMC article. - Inference of kinship using spatial distributions of SNPs for genome-wide association studies.
Lee H, Chen L. Lee H, et al. BMC Genomics. 2016 May 20;17:372. doi: 10.1186/s12864-016-2696-0. BMC Genomics. 2016. PMID: 27206321 Free PMC article. - Selecting SNPs informative for African, American Indian and European Ancestry: application to the Family Investigation of Nephropathy and Diabetes (FIND).
Williams RC, Elston RC, Kumar P, Knowler WC, Abboud HE, Adler S, Bowden DW, Divers J, Freedman BI, Igo RP Jr, Ipp E, Iyengar SK, Kimmel PL, Klag MJ, Kohn O, Langefeld CD, Leehey DJ, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Rotter JI, Schelling JR, Sedor JR, Shah VO, Smith MW, Taylor KD, Thameem F, Thornley-Brown D, Winkler CA, Guo X, Zager P, Hanson RL; FIND Research Group. Williams RC, et al. BMC Genomics. 2016 May 4;17:325. doi: 10.1186/s12864-016-2654-x. BMC Genomics. 2016. PMID: 27142425 Free PMC article. - New approaches to disease mapping in admixed populations.
Seldin MF, Pasaniuc B, Price AL. Seldin MF, et al. Nat Rev Genet. 2011 Jun 28;12(8):523-8. doi: 10.1038/nrg3002. Nat Rev Genet. 2011. PMID: 21709689 Free PMC article. Review.
Cited by
- Prospective association of a genetic risk score and lifestyle intervention with cardiovascular morbidity and mortality among individuals with type 2 diabetes: the Look AHEAD randomised controlled trial.
Look AHEAD Research Group. Look AHEAD Research Group. Diabetologia. 2015 Aug;58(8):1803-13. doi: 10.1007/s00125-015-3610-z. Epub 2015 May 14. Diabetologia. 2015. PMID: 25972230 Free PMC article. Clinical Trial. - Family-Based Rare Variant Association Analysis: A Fast and Efficient Method of Multivariate Phenotype Association Analysis.
Wang L, Lee S, Gim J, Qiao D, Cho M, Elston RC, Silverman EK, Won S. Wang L, et al. Genet Epidemiol. 2016 Sep;40(6):502-11. doi: 10.1002/gepi.21985. Epub 2016 Jun 17. Genet Epidemiol. 2016. PMID: 27312886 Free PMC article. - Extensions of BLUP Models for Genomic Prediction in Heterogeneous Populations: Application in a Diverse Switchgrass Sample.
Ramstein GP, Casler MD. Ramstein GP, et al. G3 (Bethesda). 2019 Mar 7;9(3):789-805. doi: 10.1534/g3.118.200969. G3 (Bethesda). 2019. PMID: 30651285 Free PMC article. - Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations.
Tambets K, Yunusbayev B, Hudjashov G, Ilumäe AM, Rootsi S, Honkola T, Vesakoski O, Atkinson Q, Skoglund P, Kushniarevich A, Litvinov S, Reidla M, Metspalu E, Saag L, Rantanen T, Karmin M, Parik J, Zhadanov SI, Gubina M, Damba LD, Bermisheva M, Reisberg T, Dibirova K, Evseeva I, Nelis M, Klovins J, Metspalu A, Esko T, Balanovsky O, Balanovska E, Khusnutdinova EK, Osipova LP, Voevoda M, Villems R, Kivisild T, Metspalu M. Tambets K, et al. Genome Biol. 2018 Sep 21;19(1):139. doi: 10.1186/s13059-018-1522-1. Genome Biol. 2018. PMID: 30241495 Free PMC article. - The National Longitudinal Study of Adolescent to Adult Health (Add Health) sibling pairs genome-wide data.
McQueen MB, Boardman JD, Domingue BW, Smolen A, Tabor J, Killeya-Jones L, Halpern CT, Whitsel EA, Harris KM. McQueen MB, et al. Behav Genet. 2015 Jan;45(1):12-23. doi: 10.1007/s10519-014-9692-4. Epub 2014 Nov 7. Behav Genet. 2015. PMID: 25378290 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- 5K07CA136969/CA/NCI NIH HHS/United States
- N01WH42129-32/WH/WHI NIH HHS/United States
- GM073059/GM/NIGMS NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- N01WH32100-2/WH/WHI NIH HHS/United States
- N01WH42107-26/WH/WHI NIH HHS/United States
- N01WH32122/WH/WHI NIH HHS/United States
- N01WH32105-6/WH/WHI NIH HHS/United States
- N01WH32111-13/WH/WHI NIH HHS/United States
- K07 CA136969/CA/NCI NIH HHS/United States
- N01WH24152/WH/WHI NIH HHS/United States
- R25 CA112355/CA/NCI NIH HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- N01WH32108-9/WH/WHI NIH HHS/United States
- R01 GM073059/GM/NIGMS NIH HHS/United States
- N01WH32118-32119/WH/WHI NIH HHS/United States
- K01 CA148958/CA/NCI NIH HHS/United States
- N01WH32115/WH/WHI NIH HHS/United States
- N01WH44221/WH/WHI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources