Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family - PubMed (original) (raw)
Comparative Study
. 2012 Jul 24;109(30):E2042-9.
doi: 10.1073/pnas.1205128109. Epub 2012 Jul 2.
Affiliations
- PMID: 22753491
- PMCID: PMC3409749
- DOI: 10.1073/pnas.1205128109
Comparative Study
Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family
Roswitha Krick et al. Proc Natl Acad Sci U S A. 2012.
Abstract
β-propellers that bind polyphosphoinositides (PROPPINs), a eukaryotic WD-40 motif-containing protein family, bind via their predicted β-propeller fold the polyphosphoinositides PtdIns3P and PtdIns(3,5)P(2) using a conserved FRRG motif. PROPPINs play a key role in macroautophagy in addition to other functions. We present the 3.0-Å crystal structure of Kluyveromyces lactis Hsv2, which shares significant sequence homologies with its three Saccharomyces cerevisiae homologs Atg18, Atg21, and Hsv2. It adopts a seven-bladed β-propeller fold with a rare nonvelcro propeller closure. Remarkably, in the crystal structure, the two arginines of the FRRG motif are part of two distinct basic pockets formed by a set of highly conserved residues. In comprehensive in vivo and in vitro studies of ScAtg18 and ScHsv2, we define within the two pockets a set of conserved residues essential for normal membrane association, phosphoinositide binding, and biological activities. Our experiments show that PROPPINs contain two individual phosphoinositide binding sites. Based on docking studies, we propose a model for phosphoinositide binding of PROPPINs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Structure of KlHsv2. (A) KlHsv2 forms a seven-bladed β-propeller. (B) Two sulfate ions are bound right and left of the FRRG motif (magenta). (C) Electrostatic surface potential of the molecule in the same orientation as in B. (D) Conservation of KlHsv2 is shown, based on a sequence alignment of the Hsv2 homologs from K. lactis, S. cerevisiae, Ashbya gossypii, Candida glabrata, and Yarrowia lipolytica. The dark color corresponds to a high level of the conservation. Sequences were aligned with T-Coffee (50) and analyzed with the AMAS Server (51). (E) Sulfate binding site 1. Yellow residues were selected for mutagenesis studies. Polar contacts of the sulfate ion are shown, and the distances are given in angstroms. (F) Close-up view of sulfate site 1 with the overlaid 3.0-Å resolution 2mFo-DFc electron density map contoured at 1.0 σ (blue) and the difference (mFo-DFc) omit map at +3.0 σ (green). (G) Sulfate binding site 2. Cyan-colored residues were mutated to alanines. The side chains of K245, K283, and R105 were disordered (indicated by an asterisk), and these residues were modeled as alanines. R105 is localized opposite to both sulfate binding sites near the third sulfate ion and is colored magenta. (H) Close-up view of sulfate site 2 with the overlaid electron density map (blue) and the omit difference map contoured at +3.0 σ (green). This figure was prepared with PyMOL (52).
Fig. 2.
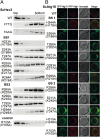
Liposome flotation assays with ScHsv2 mutants and fluorescence microscopy analysis of ScAtg18 mutants. (A) ScHsv2 mutants were added to liposomes consisting of 2% PtdIns3P, 73% phosphatidylcholine, 23% phosphatidylethanolamine, and 2% Texas Red–phosphatidylethanolamine and applied onto a Nycodenz (Progen) gradient. ScHsv2 protein was detected by immunoblotting with an anti-ScHsv2 antibody. For each ScHsv2 mutant, the corresponding KlHsv2 mutant is shown below in parentheses. (B) Fluorescence microscopic analysis of the membrane association of ScAtg18 mutants. The _atg18_Δ cells expressing the indicated GFP-ScAtg18 variants and RFP-Atg8 were grown to stationary phase in selective medium containing 0.3 mM methionine. Pictures were taken using a DeltaVision Spectris fluorescence microscope and deconvoluted using WoRx (Applied Precision) software. For each ScAtg18 mutant, the corresponding KlHsv2 mutant is shown below in parentheses.
Fig. 3.
Reflectometric interference spectroscopy and ITC measurements with ScHsv2. (A) Quantitative measurement of PtdIns3P binding of GST-ScHsv2 by reflectometric interference spectroscopy. Silicon wafers coated with a lipid bilayer consisting of DOPC and 3% PtdIns3P were incubated with increasing concentrations of GST-ScHsv2 WT protein. Binding of the protein was detected spectroscopically. The baseline was recorded in buffer. 1, formation of the DOPC bilayer [DOPC/PtdIns(3)P 97/3]; 2, rinsing with buffer; 3, stepwise addition of GST-ScHsv2 in increasing concentration; 4, rinsing with buffer. (B) Adsorption isotherm of binding of WT GST-ScHsv2 to PtdIns(3)P. A _K_d value of 1.3 ± 0.2 μM was determined via a Langmuir-fit (black line). Measurements were done in triplicate. (C) Reflectometric interference spectroscopy measurements of GST-ScHsv2 mutants. After bilayer formation, each mutant protein was added in increasing concentrations (up to 5 μM) with pauses of 5 min in between. The GST-Hsv2 mutants showed no significant membrane binding. ΔOT, changes in optical thickness (nm). (D) For ITC measurements, ScHsv2 was titrated into 2% PtdIns3P containing liposomes. The integrated areas normalized to the amount of ScHsv2 injected (kcal/mol) vs. its molar ratio to PtdIns3P are shown. Inside is a figure showing the base line-corrected raw data with power plotted against time during the injections.
Fig. 4.
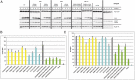
Analysis of the functional relevance of a set of conserved residues for autophagy. Cells deleted for ATG18 and cotransformed with pRS316-PGK1-GFP and pRS313-Atg18-HA, the relevant Atg18-HA mutants, or an empty plasmid as a negative control were grown to stationary phase in selection medium and shifted for 6 h to SD-(N) medium to induce macroautophagy. Samples are taken every 2 h. Only Western blots of representative strains are shown; residual blots are shown in
Fig. S6
. (A) (Top) Pgk1-GFP and GFP are detected using a GFP antibody; the amount of free GFP represents the autophagic rate. (Middle) Samples were reprobed to follow the Cvt pathway using an Ape1 antibody, which detects pApe1 and mApe1. (Bottom) Samples were further reprobed with an HA antibody to demonstrate the stability of the Atg18-HA proteins. (B) Autophagic rate is determined by quantifying the amount of free GFP using AIDA software (Raytest). The amount of free GFP of the Atg18-HA WT at 6 h is set to 100%. (C) To monitor the Cvt pathway, the percentage of mApe1 has been calculated from the total amount of pApe1 and mApe1 before starvation using AIDA software. Mutants in binding site 1 are labeled yellow, mutants of binding site 2 are shown in cyan, and mutants carrying mutations in both binding sites are shown in green. The WT, the negative control with empty plasmid, and the control mutant (K102A) with a mutation opposite to the binding sites are labeled in gray.
Fig. 5.
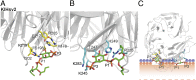
Docking of PtdIns3P into KlHsv2 and model for membrane binding of PROPPINs. Docking of PtdIns3P into binding site 1 (A) and docking into site 2 (B). Residues important for PIP binding in binding site 1 are shown in yellow and in cyan for site 2. (C) Model for membrane recognition of PROPPINs. The head groups of the two PtdIns3P molecules are shown as green sticks. The hydrophobic tails were omitted for clarity. Residues involved in PIP binding of site 1 are represented with yellow sticks, and those involved in PIP binding of site 2 are shown in cyan. The positions of the phosphates of the membrane phospholipids are shown through orange circles, whereas the violet circles represent the polar groups of these phospholipids. The shaded orange bar depicts the length of the phospholipid fatty acid chains from one leaflet of the bilayer, and the dashed orange line marks the center of the bilayer. The model was drawn approximately to scale. Loops between β-strands 6C and 6D and 7C and 7D are likely to insert into the membrane.
Fig. P1.
Membrane binding of PROPPINs. We determined the crystal structure of PROPPIN Hsv2 from the yeast K. lactis (gray). Our experiments show that PROPPINs possess two phosphoinositide binding sites and that both are required for membrane association. Based on docking studies, we propose a model for membrane association of PROPPINs in which Hsv2 binds two PtdIns3P molecules and adopts a perpendicular orientation to the membrane.
Similar articles
- It takes two to tango: PROPPINs use two phosphoinositide-binding sites.
Thumm M, Busse RA, Scacioc A, Stephan M, Janshoff A, Kühnel K, Krick R. Thumm M, et al. Autophagy. 2013 Jan;9(1):106-7. doi: 10.4161/auto.22400. Epub 2012 Oct 15. Autophagy. 2013. PMID: 23069643 Free PMC article. - How Atg18 and the WIPIs sense phosphatidylinositol 3-phosphate.
Baskaran S, Ragusa MJ, Hurley JH. Baskaran S, et al. Autophagy. 2012 Dec;8(12):1851-2. doi: 10.4161/auto.22077. Epub 2012 Sep 20. Autophagy. 2012. PMID: 22996041 Free PMC article. - Characterization of PROPPIN-Phosphoinositide Binding and Role of Loop 6CD in PROPPIN-Membrane Binding.
Busse RA, Scacioc A, Krick R, Pérez-Lara Á, Thumm M, Kühnel K. Busse RA, et al. Biophys J. 2015 May 5;108(9):2223-34. doi: 10.1016/j.bpj.2015.03.045. Biophys J. 2015. PMID: 25954880 Free PMC article. - Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18.
Watanabe Y, Kobayashi T, Yamamoto H, Hoshida H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Watanabe Y, et al. J Biol Chem. 2012 Sep 14;287(38):31681-90. doi: 10.1074/jbc.M112.397570. Epub 2012 Jul 31. J Biol Chem. 2012. PMID: 22851171 Free PMC article. - WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases.
Li D, Roberts R. Li D, et al. Cell Mol Life Sci. 2001 Dec;58(14):2085-97. doi: 10.1007/pl00000838. Cell Mol Life Sci. 2001. PMID: 11814058 Free PMC article. Review.
Cited by
- Mode of Ezrin-Membrane Interaction as a Function of PIP2 Binding and Pseudophosphorylation.
Shabardina V, Kramer C, Gerdes B, Braunger J, Cordes A, Schäfer J, Mey I, Grill D, Gerke V, Steinem C. Shabardina V, et al. Biophys J. 2016 Jun 21;110(12):2710-2719. doi: 10.1016/j.bpj.2016.05.009. Biophys J. 2016. PMID: 27332129 Free PMC article. - Cooperativity of membrane-protein and protein-protein interactions control membrane remodeling by epsin 1 and affects clathrin-mediated endocytosis.
Kroppen B, Teske N, Yambire KF, Denkert N, Mukherjee I, Tarasenko D, Jaipuria G, Zweckstetter M, Milosevic I, Steinem C, Meinecke M. Kroppen B, et al. Cell Mol Life Sci. 2021 Mar;78(5):2355-2370. doi: 10.1007/s00018-020-03647-z. Epub 2020 Sep 30. Cell Mol Life Sci. 2021. PMID: 32997199 Free PMC article. - A Dictyostelium model for BPAN disease reveals a functional relationship between the WDR45/WIPI4 homolog Wdr45l and Vmp1 in the regulation of autophagy-associated PtdIns3P and ER stress.
Tornero-Écija A, Tábara LC, Bueno-Arribas M, Antón-Esteban L, Navarro-Gómez C, Sánchez I, Vincent O, Escalante R. Tornero-Écija A, et al. Autophagy. 2022 Mar;18(3):661-677. doi: 10.1080/15548627.2021.1953262. Epub 2021 Jul 27. Autophagy. 2022. PMID: 34328055 Free PMC article. - Fluorinated Aromatic Amino Acids Distinguish Cation-π Interactions from Membrane Insertion.
He T, Gershenson A, Eyles SJ, Lee YJ, Liu WR, Wang J, Gao J, Roberts MF. He T, et al. J Biol Chem. 2015 Jul 31;290(31):19334-42. doi: 10.1074/jbc.M115.668343. Epub 2015 Jun 19. J Biol Chem. 2015. PMID: 26092728 Free PMC article. - Insights into autophagosome biogenesis from in vitro reconstitutions.
Turco E, Martens S. Turco E, et al. J Struct Biol. 2016 Oct;196(1):29-36. doi: 10.1016/j.jsb.2016.04.005. Epub 2016 May 30. J Struct Biol. 2016. PMID: 27251905 Free PMC article. Review.
References
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. - PubMed
- Krick R, Tolstrup J, Appelles A, Henke S, Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–4638. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases