Line-tension controlled mechanism for influenza fusion - PubMed (original) (raw)
Line-tension controlled mechanism for influenza fusion
Herre Jelger Risselada et al. PLoS One. 2012.
Abstract
Our molecular simulations reveal that wild-type influenza fusion peptides are able to stabilize a highly fusogenic pre-fusion structure, i.e. a peptide bundle formed by four or more trans-membrane arranged fusion peptides. We rationalize that the lipid rim around such bundle has a non-vanishing rim energy (line-tension), which is essential to (i) stabilize the initial contact point between the fusing bilayers, i.e. the stalk, and (ii) drive its subsequent evolution. Such line-tension controlled fusion event does not proceed along the hypothesized standard stalk-hemifusion pathway. In modeled influenza fusion, single point mutations in the influenza fusion peptide either completely inhibit fusion (mutants G1V and W14A) or, intriguingly, specifically arrest fusion at a hemifusion state (mutant G1S). Our simulations demonstrate that, within a line-tension controlled fusion mechanism, these known point mutations either completely inhibit fusion by impairing the peptide's ability to stabilize the required peptide bundle (G1V and W14A) or stabilize a persistent bundle that leads to a kinetically trapped hemifusion state (G1S). In addition, our results further suggest that the recently discovered leaky fusion mutant G13A, which is known to facilitate a pronounced leakage of the target membrane prior to lipid mixing, reduces the membrane integrity by forming a 'super' bundle. Our simulations offer a new interpretation for a number of experimentally observed features of the fusion reaction mediated by the prototypical fusion protein, influenza hemagglutinin, and might bring new insights into mechanisms of other viral fusion reactions.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Standard stalk-hemifusion pathway (cross-section, side-view): The initial stalk (I) radially expands (II) forming an H-shaped hemifusion diaphragm (H-HD) after the trans-leaflets (colored yellow) meet (III).
When the H-HD ruptures a fusion pore is formed (IV).
Figure 2. A -shaped hemifusion diaphragm (-HD) which is generated by a stalk that has encircled a membrane pore.
Figure 3. Evolution of a stalk in the absence and presence of a pore.
For sake of clarity the size of the lipid headgroups is exaggerated (solvent is not shown). (A) Two apposed DOPC bilayers. A preformed stalk is not stable (see Fig. 4). (B) A stable ‘hour-glass shaped’ stalk structure formed between a DOPE and DOPC bilayer (4 µs). (C) Elongation of a stalk formed between two DOPE bilayers (4 µs). (I-III) Evolution of a stalk formed between two DOPC bilayers in the vicinity of a pore (stalk-pore complex). Elongation of the stalk, which circumvents the pore, results in the formation of a  -shaped hemifusion diaphragm (
-shaped hemifusion diaphragm ( -HD).
-HD).
Figure 4. Stalk evolution in response of removing the pore (top view on porated bilayer).
(I,II,III) Sudden removal of the pore before completion of the  -HD reverses the stalk elongation process, and the stalk completely disappears (DOPC at 310 K, with
-HD reverses the stalk elongation process, and the stalk completely disappears (DOPC at 310 K, with  water molecules per lipid between the membranes). Hydrophobic lipid tails are colored grey, polar-headgroups (DOPC) tan.
water molecules per lipid between the membranes). Hydrophobic lipid tails are colored grey, polar-headgroups (DOPC) tan.
Figure 5. (A) Overlap between the coarse-grained model (backbone red and side-chains yellow) of the wild-type influenza fusion peptide and the NMR structure .
The two helices are joined by a linker region at a slightly bent angle (boomerang-shape). (B) The wild-type influenza fusion peptides (side-chains not shown) aggregate into a stable hexameric bundle. The bundle interior is depleted in solvent (colored blue) and lipid head groups. For sake of clarity, the first backbone residue (Gly1) is colored yellow. (C) Top view of the bundle. The bundle’s interior is mainly composed of the hydrophilic residues Glu11 (colored blue) and Asn12 (colored green) that are located in the kinked region of the peptide and which point toward the central axis of the bundle.
Figure 6. Evolution of the stalk in the presence of the peptide bundle.
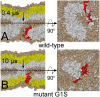
For sake of clarity the size of the lipid headgroups is exaggerated (solvent is not shown). (A) The elongated stalk (wild-type peptides) after 0.4  s. The bundle has opened up and the stalk and has partly surrounded the formed hole. Notice the readily adopted banana-shape. The stalk forces the peptides to the remaining rim portion. At this stage mixing occurs between both the _cis_-leaflets and the _trans_-leaflet of the target membrane (colored gray), while the _cis_-leaflet of the host cell (colored yellow) does not contribute to lipid mixing. (B) Mutating a single residue in the peptides, Gly1 to Ser1 (colored green), known as the terminal hemifusion mutant G1S , , stabilizes both bundle and stalk but inhibits elongation of the stalk (10
s. The bundle has opened up and the stalk and has partly surrounded the formed hole. Notice the readily adopted banana-shape. The stalk forces the peptides to the remaining rim portion. At this stage mixing occurs between both the _cis_-leaflets and the _trans_-leaflet of the target membrane (colored gray), while the _cis_-leaflet of the host cell (colored yellow) does not contribute to lipid mixing. (B) Mutating a single residue in the peptides, Gly1 to Ser1 (colored green), known as the terminal hemifusion mutant G1S , , stabilizes both bundle and stalk but inhibits elongation of the stalk (10  s). Consequentially, the fusion reaction becomes trapped. Note that lipid head-groups are excluded from the pore interior and the _trans_-leaflets (colored yellow) are hindered from participating in the lipid mixing.
s). Consequentially, the fusion reaction becomes trapped. Note that lipid head-groups are excluded from the pore interior and the _trans_-leaflets (colored yellow) are hindered from participating in the lipid mixing.
Figure 7. Point mutations that are known to inhibit fusion destabilize the bundle.
(top) Mutating a single residue, Gly1 to Val1 (colored blue), destabilizes the peptide bundle (mutant G1V). (bottom) Mutating a single residue, Trp14 to Ala14 (colored cyan), rapidly destabilizes the peptide bundle (mutant W14A). Notice the flexible kink that points out of the membrane .
Figure 8. The G1S mutation reverses the stalk elongation process facilitated by the wild-type.
Notice the removal of solvent (colored blue) and lipid head-groups (colored tan) from the membrane interior when the peptide bundle ‘reseals’ itself – the stalk and peptide are competitive lineactants.
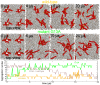
Figure 9. Interaction between multiple peptide bundles.
(upper panel) Four wild-type bundles (top-view). The bundles strongly repel each other and maximize their separation distance in the course of the simulation. Eventually one of the bundles vanishes. (middle panel) Four G13A mutant bundles. The bundles are attractive and their coalescence results in a ‘super’ bundle consisting of 10 trans-membrane arranged peptides. (lower panel) Aggregation number of the largest bundle in the course of the simulation (Only the trans-membrane arranged peptide are counted). The brown line shows a separate simulation where the G13A mutation is reversed after 20  s (G13A -> wild-type). The wild-type ‘super’ bundle readopts its usual size in the course of the simulation.
s (G13A -> wild-type). The wild-type ‘super’ bundle readopts its usual size in the course of the simulation.
Figure 10. Detailed view of the ‘super’ bundle (10 s) formed by the leaky fusion mutant G13A (Top view, cross-section through the bilayer center).
Notice that residue 13 (colored green) directly faces the hydrophobic lipid rim around the bundle. The solvent (colored blue) in the center of the bundle suggests the occurrence of leakage prior to lipid mixing .
Similar articles
- Single mutation effects on conformational change and membrane deformation of influenza hemagglutinin fusion peptides.
Li J, Das P, Zhou R. Li J, et al. J Phys Chem B. 2010 Jul 8;114(26):8799-806. doi: 10.1021/jp1029163. J Phys Chem B. 2010. PMID: 20552971 - Hemagglutinin fusion peptide mutants in model membranes: structural properties, membrane physical properties, and PEG-mediated fusion.
Haque ME, Chakraborty H, Koklic T, Komatsu H, Axelsen PH, Lentz BR. Haque ME, et al. Biophys J. 2011 Sep 7;101(5):1095-104. doi: 10.1016/j.bpj.2011.07.031. Biophys J. 2011. PMID: 21889446 Free PMC article. - Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion.
Hamilton BS, Whittaker GR, Daniel S. Hamilton BS, et al. Viruses. 2012 Jul;4(7):1144-68. doi: 10.3390/v4071144. Epub 2012 Jul 24. Viruses. 2012. PMID: 22852045 Free PMC article. Review. - Architecture of the influenza hemagglutinin membrane fusion site.
Bentz J, Mittal A. Bentz J, et al. Biochim Biophys Acta. 2003 Jul 11;1614(1):24-35. doi: 10.1016/s0005-2736(03)00160-3. Biochim Biophys Acta. 2003. PMID: 12873763 Review.
Cited by
- Optimization of an elastic network augmented coarse grained model to study CCMV capsid deformation.
Globisch C, Krishnamani V, Deserno M, Peter C. Globisch C, et al. PLoS One. 2013 Apr 16;8(4):e60582. doi: 10.1371/journal.pone.0060582. Print 2013. PLoS One. 2013. PMID: 23613730 Free PMC article. - Molecular mechanisms of the influenza fusion peptide: insights from experimental and simulation studies.
Lousa D, Soares CM. Lousa D, et al. FEBS Open Bio. 2021 Dec;11(12):3253-3261. doi: 10.1002/2211-5463.13323. Epub 2021 Nov 8. FEBS Open Bio. 2021. PMID: 34710289 Free PMC article. Review. - How proteins open fusion pores: insights from molecular simulations.
Risselada HJ, Grubmüller H. Risselada HJ, et al. Eur Biophys J. 2021 Mar;50(2):279-293. doi: 10.1007/s00249-020-01484-3. Epub 2020 Dec 19. Eur Biophys J. 2021. PMID: 33340336 Free PMC article. Review. - Characterization of Lipid-Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation.
Muller MP, Jiang T, Sun C, Lihan M, Pant S, Mahinthichaichan P, Trifan A, Tajkhorshid E. Muller MP, et al. Chem Rev. 2019 May 8;119(9):6086-6161. doi: 10.1021/acs.chemrev.8b00608. Epub 2019 Apr 12. Chem Rev. 2019. PMID: 30978005 Free PMC article. Review. - Fusing simulation and experiment: The effect of mutations on the structure and activity of the influenza fusion peptide.
Lousa D, Pinto AR, Victor BL, Laio A, Veiga AS, Castanho MA, Soares CM. Lousa D, et al. Sci Rep. 2016 Jun 15;6:28099. doi: 10.1038/srep28099. Sci Rep. 2016. PMID: 27302370 Free PMC article.
References
- Kozlov MM, Markin VS. Possible mechanism of membrane fusion. Biofizika. 1983;28:255–261. - PubMed
- Schick M. Membrane fusion; the emergence of a new paradigm. J Stat Phys. 2011;142:1317.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical