New readers and interpretations of poly(ADP-ribosyl)ation - PubMed (original) (raw)
Review
New readers and interpretations of poly(ADP-ribosyl)ation
Thomas Kalisch et al. Trends Biochem Sci. 2012 Sep.
Abstract
Poly(ADP-ribosyl)ation (PARylation), a protein post-translational modification that was originally connected to the DNA damage response, is now known to engage in a continuously increasing number of biological processes. Despite extensive research and ceaseless, important findings about its role and mode of action, poly(ADP-ribose) remains an enigma regarding its structural complexity and diversity. The recent identification and structural characterization of four different poly(ADP-ribose) binding motifs represents a quantum leap in the comprehension of how this molecule can be decoded. Moreover, the recent discovery of a direct connection between PARylation and poly-ubiquitylation in targeting proteins for degradation by the proteasome has paved the way for a new interpretation of this protein modification. These two novel aspects, poly(ADP-ribose) recognition and readout by the ubiquitylation/proteasome system are developed here.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
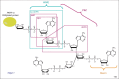
Figure 1
Structure of poly(ADP-ribose) (PAR) and recognition by PAR-binding motifs (PBMs). The chemical structure of branched PAR, covalently attached to an acceptor protein (which can be poly(ADP-ribose) polymerase itself), is shown. The characteristic linkages of linear (1→2′ O-glycosidic bound) and branched (1→2′′ O-glycosidic bound) are indicated. The brackets illustrate the region recognized by each PAR-binding domain. The ADP-ribose (ADPR) derivatives used in structural characterization of PAR-binding domains are framed. The RNF146 PAR-binding domain WWE was co-crystallized with iso-ADPR , supporting a mechanism of internal recognition of PAR. The solution structure of the two aprataxin and PNK-like factor (APLF) PAR-binding zinc fingers (PBZ motifs) bound to 2′-O-α-D-ribofuranosyl-adenosine (RFA) was solved by nuclear magnetic resonance (NMR) , whereas the CHFR PBZ domain was co-crystallized with AMP and ADPR . This PBZ domain was also co-crystallized with P1P2-diadenosine 5′-pyrophosphate (AMP2), which is not a derivative of PAR and thus cannot be illustrated in this scheme, but which supports the hypothesis that the PBZ motif recognizes two successive ADPR units. Macrodomains bind ADPR units, but capping of the 2′OH from ADP prohibits internal recognition of PAR , . How the PBMs recognize PAR remains unknown, as illustrated by the open question mark.
Figure 2
Schematic domain architecture of macro-, poly(ADP-ribose)-binding zinc finger (PBZ) and WWE domain containing human proteins. Protein domains illustrated by colored boxes are defined according to the Pfam 26.0 database. (a) Schematic representation of human macrodomain (blue box) containing proteins. Their ability to bind poly(ADP-ribose) (PAR), ADP-ribose (ADPR), or O-acetyl-ADP-ribose (OAADPR), and their hydrolyzing activity toward ADPR-1′′P or OAADPR is indicated, if known. Structural information, when available in the protein data bank (PDB), is indicated. (b) Schematic representation of human PBZ domain containing proteins. Their ability to bind PAR (if known), and the availability of their 3D structures, are indicated. (c) Schematic representation of human WWE domain containing proteins. These WWE-containing proteins are classified according to whether their associated domains are either endowed with poly(ADP-ribose) polymerase (PARP) activity or with ubiquitin E3 ligase activity. Also indicated is whether these WWE domains possess the ability to bind PAR or isoADPR (if known), and whether 3D structural information is available. DDHD, Asp- and His-containing motif involved in phospholipase activity (light gray box); H2A, domain homologous to histone H2A (pale violet box); helicase-C, helicase superfamily C-terminal domain associated with DEXDc-, DEAD- and DEAH-box proteins (lavender box); FHA, forkhead associated (light pink box); HECT, homologous to E6-AP carboxyl terminus domain, displaying E3-ligase activity (light orange box); lactamase B, domain homologous to β-lactamase endowed with nuclease activity (gray box); macro, homologous to the nonhistone part of macroH2A, displaying PAR-binding activity (see text; blue box); PARP, catalytic domain, homologous to the poly(ADP-ribose) synthesis domain of PARP-1, endowed with mono- or poly(ADP-ribosyl)ation activity (green box); PBZ, PAR-binding zinc finger (see text; orange box); RING, really interesting new gene: zinc binding domain with ubiquitin E3 ligase activity (sienna box); SAM-L, sterile α motif-like (purple box); SNF2-N, SNF2 family N-terminal domain (light steel blue box); UBA, ubiquitin-binding domain (silver gray box); WWE, named after its three conserved residues Trp, Trp and Glu, displaying PAR-binding activity (see text; dark yellow box); Zf-CCCH, zinc finger motif (pink box).
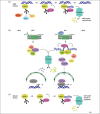
Figure 3
poly(ADP-ribose) (PAR)-dependent poly-ubiquitylation of proteins to promote their degradation by the proteasome. (a) PAR-dependent activation of RNF146 by poly(ADP-ribose) polymerase (PARP)-1 in the DNA damage response. PARP-1 detects DNA breaks, is activated, and auto- or heteromodifies acceptor proteins. PAR produced at DNA damage site has a signaling role and directly recruits factors involved in regulation of chromatin structure and DNA repair [for example amplified in liver cancer 1 (ALC1), X-ray repair cross-complementing gene 1 (XRCC1) or meiotic recombination 11 (Mre11), see Box 1]. The ubiquitin E3 ligase RNF146 is activated upon binding to poly(ADP-ribosyl)ated (PARylated) PARP-1 and poly-ubiquitylates PARP-1. Ubiquitylated PARP-1 is subsequently targeted to the proteasome for degradation. The timely and orchestrated poly(ADP-ribosyl)ation and ubiquitylation of PARP-1 regulates DNA repair and favors cell survival. (b) PAR-dependent activation of RNF146 by tankyrases during Wnt signaling. In the absence of Wnt, the multiprotein β-catenin destruction complex, which contains adenomatous polyposis coli (APC), glycogen synthase kinase 3 (GSK3) and axin triggers the proteasome-dependent degradation of β-catenin. The Wnt signaling cascade is activated by binding of Wnt to its receptor, the low-density lipoprotein receptor-related protein LRP/Frizzled. Tankyrase 1 and 2 (TNKS1/2) PARylate themselves and axin. How TNKS1/2 are activated during Wnt signaling is unknown, and it remains to be determined by which mechanism the activation signal is transmitted from Wnt receptor to TNKS1/2 (illustrated by a dashed arrow between the Wnt receptor and TNKS1/2/RNF146 complex). The PAR activates the ubiquitin E3 ligase RNF146, leading to TNKS1/2 and axin poly-ubiquitylation and subsequent degradation by the proteasome. The multiprotein β-catenin destruction complex is destabilized by the absence of axin, leading to β-catenin accumulation and translocation into the nucleus, where it coactivates the T-cell factor/lymphoid enhancer (TCF/LEF) transcription factor, to promote transcription of Wnt-dependent genes. (c) PAR-dependent activation of CHFR [checkpoint with forkhead (FHA)-associated and really interesting new gene (RING) finger domains] by PARP-1 during mitotic stress. In response to mitotic stress caused by drugs that affect microtubules, such as nocodazole or docetaxel, PARP-1 is activated by an as-yet-unknown process, leading to its auto-PARylation. This results in the PAR-dependent activation of the ubiquitin E3 ligase CHFR and subsequent poly-ubiquitylation of PARP-1 and degradation by the proteasome. Removal of PARylated PARP-1 is necessary to promote cell cycle arrest at prophase.
Similar articles
- Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions.
D'Amours D, Desnoyers S, D'Silva I, Poirier GG. D'Amours D, et al. Biochem J. 1999 Sep 1;342 ( Pt 2)(Pt 2):249-68. Biochem J. 1999. PMID: 10455009 Free PMC article. Review. - Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins.
Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Ahel I, et al. Nature. 2008 Jan 3;451(7174):81-5. doi: 10.1038/nature06420. Nature. 2008. PMID: 18172500 - Poly(ADP-ribose): PARadigms and PARadoxes.
Bürkle A, Virág L. Bürkle A, et al. Mol Aspects Med. 2013 Dec;34(6):1046-65. doi: 10.1016/j.mam.2012.12.010. Epub 2013 Jan 2. Mol Aspects Med. 2013. PMID: 23290998 Review. - Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Wang Z, et al. Genes Dev. 2012 Feb 1;26(3):235-40. doi: 10.1101/gad.182618.111. Epub 2012 Jan 19. Genes Dev. 2012. PMID: 22267412 Free PMC article. - 50Years of poly(ADP-ribosyl)ation.
Virág L. Virág L. Mol Aspects Med. 2013 Dec;34(6):1043-5. doi: 10.1016/j.mam.2013.05.002. Epub 2013 May 28. Mol Aspects Med. 2013. PMID: 23727362 Review.
Cited by
- A novel role for the mono-ADP-ribosyltransferase PARP14/ARTD8 in promoting homologous recombination and protecting against replication stress.
Nicolae CM, Aho ER, Choe KN, Constantin D, Hu HJ, Lee D, Myung K, Moldovan GL. Nicolae CM, et al. Nucleic Acids Res. 2015 Mar 31;43(6):3143-53. doi: 10.1093/nar/gkv147. Epub 2015 Mar 9. Nucleic Acids Res. 2015. PMID: 25753673 Free PMC article. - PARP10 promotes cellular proliferation and tumorigenesis by alleviating replication stress.
Schleicher EM, Galvan AM, Imamura-Kawasawa Y, Moldovan GL, Nicolae CM. Schleicher EM, et al. Nucleic Acids Res. 2018 Sep 28;46(17):8908-8916. doi: 10.1093/nar/gky658. Nucleic Acids Res. 2018. PMID: 30032250 Free PMC article. - Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation.
Feijs KL, Forst AH, Verheugd P, Lüscher B. Feijs KL, et al. Nat Rev Mol Cell Biol. 2013 Jul;14(7):443-51. doi: 10.1038/nrm3601. Epub 2013 Jun 5. Nat Rev Mol Cell Biol. 2013. PMID: 23736681 Free PMC article. Review. - Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair.
Zhao Y, Brickner JR, Majid MC, Mosammaparast N. Zhao Y, et al. Trends Cell Biol. 2014 Jul;24(7):426-34. doi: 10.1016/j.tcb.2014.01.005. Epub 2014 Feb 23. Trends Cell Biol. 2014. PMID: 24569222 Free PMC article. Review. - Autophagy requires poly(adp-ribosyl)ation-dependent AMPK nuclear export.
Rodríguez-Vargas JM, Rodríguez MI, Majuelos-Melguizo J, García-Diaz Á, González-Flores A, López-Rivas A, Virág L, Illuzzi G, Schreiber V, Dantzer F, Oliver FJ. Rodríguez-Vargas JM, et al. Cell Death Differ. 2016 Dec;23(12):2007-2018. doi: 10.1038/cdd.2016.80. Epub 2016 Sep 30. Cell Death Differ. 2016. PMID: 27689873 Free PMC article.
References
- Hottiger M.O. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. - PubMed
- Kleine H. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous