3D Slicer as an image computing platform for the Quantitative Imaging Network - PubMed (original) (raw)
. 2012 Nov;30(9):1323-41.
doi: 10.1016/j.mri.2012.05.001. Epub 2012 Jul 6.
Reinhard Beichel, Jayashree Kalpathy-Cramer, Julien Finet, Jean-Christophe Fillion-Robin, Sonia Pujol, Christian Bauer, Dominique Jennings, Fiona Fennessy, Milan Sonka, John Buatti, Stephen Aylward, James V Miller, Steve Pieper, Ron Kikinis
Affiliations
- PMID: 22770690
- PMCID: PMC3466397
- DOI: 10.1016/j.mri.2012.05.001
3D Slicer as an image computing platform for the Quantitative Imaging Network
Andriy Fedorov et al. Magn Reson Imaging. 2012 Nov.
Abstract
Quantitative analysis has tremendous but mostly unrealized potential in healthcare to support objective and accurate interpretation of the clinical imaging. In 2008, the National Cancer Institute began building the Quantitative Imaging Network (QIN) initiative with the goal of advancing quantitative imaging in the context of personalized therapy and evaluation of treatment response. Computerized analysis is an important component contributing to reproducibility and efficiency of the quantitative imaging techniques. The success of quantitative imaging is contingent on robust analysis methods and software tools to bring these methods from bench to bedside. 3D Slicer is a free open-source software application for medical image computing. As a clinical research tool, 3D Slicer is similar to a radiology workstation that supports versatile visualizations but also provides advanced functionality such as automated segmentation and registration for a variety of application domains. Unlike a typical radiology workstation, 3D Slicer is free and is not tied to specific hardware. As a programming platform, 3D Slicer facilitates translation and evaluation of the new quantitative methods by allowing the biomedical researcher to focus on the implementation of the algorithm and providing abstractions for the common tasks of data communication, visualization and user interface development. Compared to other tools that provide aspects of this functionality, 3D Slicer is fully open source and can be readily extended and redistributed. In addition, 3D Slicer is designed to facilitate the development of new functionality in the form of 3D Slicer extensions. In this paper, we present an overview of 3D Slicer as a platform for prototyping, development and evaluation of image analysis tools for clinical research applications. To illustrate the utility of the platform in the scope of QIN, we discuss several use cases of 3D Slicer by the existing QIN teams, and we elaborate on the future directions that can further facilitate development and validation of imaging biomarkers using 3D Slicer.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1
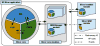
3D Slicer “ecosystem”. Slicer is a cross-platform package, but certain requirements on the graphics system, such as OpenGL drivers, should be met to support accelerated rendering. Its dependency libraries are portable across platforms and are distributed under compatible licenses. Those dependencies that are part of NA-MIC Kit often share the 3D Slicer developer community and are developed in synergy with the Slicer efforts. Slicer itself consists of the main application framework (core) and plugins (modules). Custom functionality is introduced by implementing external modules (Slicer extensions).
Figure 2
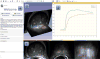
Experimental visualization of dynamic contrast enhancement (DCE) MRI of the prostate demonstrating various visualization capabilities of 3D Slicer. Slice viewers (annotated as “1”) show an axial cross-section of T2w image, and a single frame of a DCE series in axial and coronal cross-sections. Contours of the segmented areas corresponding to normal-appearing and tumor-suspected areas appear in colored overlay. Chart viewer shows the mean change in signal intensity during DCE imaging for the contoured areas (annotated as “2”), while the 3D viewer (annotated as “3”) brings cross-sections and segmented areas (represented by triangulated surfaces) into 3D context. Panel on the left shows the GUI of the Slicer Welcome module (annotated as “4”).
Figure 3
High-level view of the 3D Slicer architecture. Slicer core consists of the GUI, Logic and MRML components that follow the MVC design pattern (MRML is implemented as a separate library). The same pattern is replicated in the loadable modules to promote separation of concerns in the code. Communication between the individual elements is enabled by API calls and event mechanisms. SEM modules communicate with the application via the memory and/or file IO, mediated by a designated core module, and do not have GUI/View components they can directly or interactively manipulate.
Figure 4
Visualization capabilities and Slicer user interface are utilized for exploration of the multiparametric MRI of the prostate. The T2-weighted (T2w) MR scans (top left image and the bottom row of consecutive slices) provide anatomical detail and context for interpretation of the registered Apparent Diffusion Coefficient (ADC) map (first row of the image matrix) calculated from the Diffusion Weighted MRI (DWI) and various pharmacokinetic maps computed from dynamic contrast enhanced (DCE) MRI, as shown in the 2D axial slice viewer (marked by “1”) and the Compare View layout below. Spatial context of the annotated tumor ROI is provided by composed view of the tumor ROI surface and the T2w image cross-section in the 3D viewer (marked by “2”).
Figure 5
Iowa-developed integrated Slicer module for tumor uptake quantification. Slice viewers show cross-sections of a PET/CT H&N cancer dataset, with the crosshairs at the tumor location. The tumor was segmented using a custom segmentation method developed for this application. In addition, a volumetric reference region was defined (see sagittal cross-section). Based on tumor and reference regions, several quantitative indices were calculated and are summarized in the window in the upper right part of the image. Note that the module was developed using 3D Slicer software version 3 (in contrast, Figures 2 and 4 illustrate the interface of the current, 4th generation of the software).
Figure 6
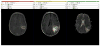
T1, FLAIR and ADC maps are registered and segmented using the grow-cut segmentation module in Slicer. In the left pane, the enhancing region is delineated on a post-contrast T1 image, while in the center the region of FLAIR abnormality is identified. In the right pane a region of vasogenic edema is outlined on the ADC map derived from the DWI images.
Similar articles
- 3D Slicer as a tool for interactive brain tumor segmentation.
Kikinis R, Pieper S. Kikinis R, et al. Annu Int Conf IEEE Eng Med Biol Soc. 2011;2011:6982-4. doi: 10.1109/IEMBS.2011.6091765. Annu Int Conf IEEE Eng Med Biol Soc. 2011. PMID: 22255945 Free PMC article. - SlicerRT: radiation therapy research toolkit for 3D Slicer.
Pinter C, Lasso A, Wang A, Jaffray D, Fichtinger G. Pinter C, et al. Med Phys. 2012 Oct;39(10):6332-8. doi: 10.1118/1.4754659. Med Phys. 2012. PMID: 23039669 - Polymorph segmentation representation for medical image computing.
Pinter C, Lasso A, Fichtinger G. Pinter C, et al. Comput Methods Programs Biomed. 2019 Apr;171:19-26. doi: 10.1016/j.cmpb.2019.02.011. Epub 2019 Feb 21. Comput Methods Programs Biomed. 2019. PMID: 30902247 - SlicerHeart: An open-source computing platform for cardiac image analysis and modeling.
Lasso A, Herz C, Nam H, Cianciulli A, Pieper S, Drouin S, Pinter C, St-Onge S, Vigil C, Ching S, Sunderland K, Fichtinger G, Kikinis R, Jolley MA. Lasso A, et al. Front Cardiovasc Med. 2022 Sep 6;9:886549. doi: 10.3389/fcvm.2022.886549. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36148054 Free PMC article. Review. - Exploring the Application of the Artificial-Intelligence-Integrated Platform 3D Slicer in Medical Imaging Education.
Zhang Y, Feng H, Zhao Y, Zhang S. Zhang Y, et al. Diagnostics (Basel). 2024 Jan 8;14(2):146. doi: 10.3390/diagnostics14020146. Diagnostics (Basel). 2024. PMID: 38248022 Free PMC article. Review.
Cited by
- Mitochondria are absent from microglial processes performing surveillance, chemotaxis, and phagocytic engulfment.
Pietramale AN, Bame X, Doty ME, Hill RA. Pietramale AN, et al. bioRxiv [Preprint]. 2024 Oct 18:2024.10.15.618505. doi: 10.1101/2024.10.15.618505. bioRxiv. 2024. PMID: 39463986 Free PMC article. Preprint. - Evaluation of a free 3D software for kidney stones' surgical planning: "kidney stone calculator" a pilot study.
Panthier F, Traxer O, Yonneau L, Lebret T, Berthe L, Illoul L, Timsit MO, Mejean A, Doizi S, Audenet F. Panthier F, et al. World J Urol. 2021 Sep;39(9):3607-3614. doi: 10.1007/s00345-021-03671-z. Epub 2021 Mar 29. World J Urol. 2021. PMID: 33779821 Free PMC article. - The association between serum S100β levels and prognosis in acute stroke patients after intravenous thrombolysis: a multicenter prospective cohort study.
Qu Y, Jin H, Abuduxukuer R, Qi S, Si XK, Zhang P, Zhang KJ, Wang SJ, Zheng XY, Zhang Y, Gao JH, Zhang XK, Liu XD, Li CY, Li GC, Wang J, Jin H, He Y, Jiang L, Liu L, Jiang Y, Teng RH, Jia Y, Zhang BJ, Chen Z, Qi Y, Liu X, Li S, Sun X, Nguyen TN, Yang Y, Guo ZN; Biomarkers of Brain Injury Investigator Study Group. Qu Y, et al. BMC Med. 2024 Oct 3;22(1):304. doi: 10.1186/s12916-024-03517-6. BMC Med. 2024. PMID: 39358745 Free PMC article. - Sensitivity analysis of transabdominal fetal pulse oximetry using MRI-based simulations.
Wu J, Satish G, Ruesch A, Jayet B, Komolibus K, Andersson-Engels S, Debreczeny MP, Kainerstorfer JM. Wu J, et al. Biomed Opt Express. 2024 Aug 19;15(9):5280-5295. doi: 10.1364/BOE.531149. eCollection 2024 Sep 1. Biomed Opt Express. 2024. PMID: 39296401 Free PMC article. - CT imaging-derived phenotypes for abdominal muscle and their association with age and sex in a medical biobank.
Vu PT, Chahine C, Chatterjee N, MacLean MT, Swago S, Bhattaru A, Thompson EW, Ikhlas A, Oteng E, Davidson L, Tran R, Hazim M, Raghupathy P, Verma A, Duda J, Gee J, Luks V, Gershuni V, Wu G, Rader D, Sagreiya H, Witschey WR; Penn Medicine Biobank. Vu PT, et al. Sci Rep. 2024 Jun 26;14(1):14807. doi: 10.1038/s41598-024-64603-6. Sci Rep. 2024. PMID: 38926479 Free PMC article.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. - PubMed
- Park JW, Kerbel RS, Kelloff GJ, Barrett JC, Chabner Ba, Parkinson DR, Peck J, Ruddon RW, Sigman CC, Slamon DJ. Rationale for biomarkers and surrogate end points in mechanism-driven oncology drug development. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004 Jun 1;10(11):3885–96. - PubMed
- Eisenhauer E, Therasse P, Bogaerts J, Schwartz L, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European journal of cancer (Oxford, England: 1990) 2009 Jan;45(2):228–247. - PubMed
- Jaffe Ta, Wickersham NW, Sullivan DC. Quantitative imaging in oncology patients: Part 1, radiology practice patterns at major U.S. cancer centers. AJR American journal of roentgenology. 2010 Jul;195(1):101–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01-CA140206/CA/NCI NIH HHS/United States
- U54EB005149/EB/NIBIB NIH HHS/United States
- U01 CA140206/CA/NCI NIH HHS/United States
- R01 EB004640/EB/NIBIB NIH HHS/United States
- U01 CA151261/CA/NCI NIH HHS/United States
- 1U01CA154601-01/CA/NCI NIH HHS/United States
- R01 CA111288/CA/NCI NIH HHS/United States
- U01 CA154601/CA/NCI NIH HHS/United States
- P41EB015898/EB/NIBIB NIH HHS/United States
- P41RR13218/RR/NCRR NIH HHS/United States
- U01CA151261/CA/NCI NIH HHS/United States
- U54 EB005149/EB/NIBIB NIH HHS/United States
- P41 RR019703/RR/NCRR NIH HHS/United States
- R00 LM009889/LM/NLM NIH HHS/United States
- P41 EB015898/EB/NIBIB NIH HHS/United States
- P41 RR013218/RR/NCRR NIH HHS/United States
- U01 CA154602/CA/NCI NIH HHS/United States
- 4R00LM009889-03/LM/NLM NIH HHS/United States
- 1R01CA111288/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials