Ultrafast clustering algorithms for metagenomic sequence analysis - PubMed (original) (raw)
Ultrafast clustering algorithms for metagenomic sequence analysis
Weizhong Li et al. Brief Bioinform. 2012 Nov.
Abstract
The rapid advances of high-throughput sequencing technologies dramatically prompted metagenomic studies of microbial communities that exist at various environments. Fundamental questions in metagenomics include the identities, composition and dynamics of microbial populations and their functions and interactions. However, the massive quantity and the comprehensive complexity of these sequence data pose tremendous challenges in data analysis. These challenges include but are not limited to ever-increasing computational demand, biased sequence sampling, sequence errors, sequence artifacts and novel sequences. Sequence clustering methods can directly answer many of the fundamental questions by grouping similar sequences into families. In addition, clustering analysis also addresses the challenges in metagenomics. Thus, a large redundant data set can be represented with a small non-redundant set, where each cluster can be represented by a single entry or a consensus. Artifacts can be rapidly detected through clustering. Errors can be identified, filtered or corrected by using consensus from sequences within clusters.
Figures
Figure 1:
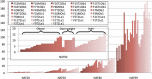
Distribution of microbial diversity measured by NATs (NAT20, NAT50, NAT80 and NAT99) for 33 human gut samples. The _x_-axis is NAT category. The _y_-axis is NAT value. Samples are colored by sample type (obese, over weight or lean). The results show that obese samples have less average NAT50 than the lean samples.
Figure 2:
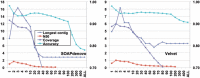
Assembly performance of the filtered reads for metagenomic sample MH0006. _x_-axis is the redundancy cutoff N. The length of the longest contig (kb) and N50 (kb) are plotted against the left _y_-axis. The accuracy and genome coverage are against the right _y_-axis. The assembly results for original reads are at far right side marked as ‘ALL’ on _x_-axis. The accuracy of contigs is the total length of correct contigs divided by the total length of all contigs. The genome coverage is the fraction of reference genome covered by the correct contigs.
Figure 3:
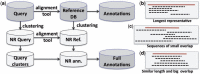
Using NR query and NR reference database for metagenome annotation.
Figure 4:
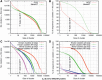
Distribution of GOS and MetaHIT protein clusters. The _x_-axis is the cluster size X. The _y_-axis in left figures is the number of clusters of size at least X; the _y_-axis in right figures is the percentage of total sequences included in the clusters of size at least X. Graphs in (A) and (B) are for all GOS and MetaHIT sequences. Graphs in (C) and (D) are only for MetaHIT sequences, grouped by Known and Novel clusters. In addition, two separate lines are made for NR sequences (i.e. the 3 076 514 representative sequences clustered at 90% identity).
Similar articles
- Analysis and comparison of very large metagenomes with fast clustering and functional annotation.
Li W. Li W. BMC Bioinformatics. 2009 Oct 28;10:359. doi: 10.1186/1471-2105-10-359. BMC Bioinformatics. 2009. PMID: 19863816 Free PMC article. - Estimating the composition of species in metagenomes by clustering of next-generation read sequences.
Seok HS, Hong W, Kim J. Seok HS, et al. Methods. 2014 Oct 1;69(3):213-9. doi: 10.1016/j.ymeth.2014.07.009. Epub 2014 Jul 27. Methods. 2014. PMID: 25072168 - OGRE: Overlap Graph-based metagenomic Read clustEring.
Balvert M, Luo X, Hauptfeld E, Schönhuth A, Dutilh BE. Balvert M, et al. Bioinformatics. 2021 May 17;37(7):905-912. doi: 10.1093/bioinformatics/btaa760. Bioinformatics. 2021. PMID: 32871010 Free PMC article. - Metagenomic Assembly: Overview, Challenges and Applications.
Ghurye JS, Cepeda-Espinoza V, Pop M. Ghurye JS, et al. Yale J Biol Med. 2016 Sep 30;89(3):353-362. eCollection 2016 Sep. Yale J Biol Med. 2016. PMID: 27698619 Free PMC article. Review. - Classification of metagenomic sequences: methods and challenges.
Mande SS, Mohammed MH, Ghosh TS. Mande SS, et al. Brief Bioinform. 2012 Nov;13(6):669-81. doi: 10.1093/bib/bbs054. Epub 2012 Sep 8. Brief Bioinform. 2012. PMID: 22962338 Review.
Cited by
- Influence of milk microbiota on Listeria monocytogenes survival during cheese ripening.
Lee J, Seo Y, Ha J, Kim S, Choi Y, Oh H, Lee Y, Kim Y, Kang J, Park E, Yoon Y. Lee J, et al. Food Sci Nutr. 2020 Jul 31;8(9):5071-5076. doi: 10.1002/fsn3.1806. eCollection 2020 Sep. Food Sci Nutr. 2020. PMID: 32994967 Free PMC article. - Influence of dietary n-3 long-chain fatty acids on microbial diversity and composition of sows' feces, colostrum, milk, and suckling piglets' feces.
Llauradó-Calero E, Climent E, Chenoll E, Ballester M, Badiola I, Lizardo R, Torrallardona D, Esteve-Garcia E, Tous N. Llauradó-Calero E, et al. Front Microbiol. 2022 Dec 5;13:982712. doi: 10.3389/fmicb.2022.982712. eCollection 2022. Front Microbiol. 2022. PMID: 36545207 Free PMC article. - Detection of bacterial DNA from central venous catheter removed from patients by next generation sequencing: a preliminary clinical study.
Okuda KI, Yoshii Y, Yamada S, Chiba A, Hironaka I, Hori S, Yanaga K, Mizunoe Y. Okuda KI, et al. Ann Clin Microbiol Antimicrob. 2018 Dec 22;17(1):44. doi: 10.1186/s12941-018-0297-2. Ann Clin Microbiol Antimicrob. 2018. PMID: 30577829 Free PMC article. - Endotoxemia by Porphyromonas gingivalis Injection Aggravates Non-alcoholic Fatty Liver Disease, Disrupts Glucose/Lipid Metabolism, and Alters Gut Microbiota in Mice.
Sasaki N, Katagiri S, Komazaki R, Watanabe K, Maekawa S, Shiba T, Udagawa S, Takeuchi Y, Ohtsu A, Kohda T, Tohara H, Miyasaka N, Hirota T, Tamari M, Izumi Y. Sasaki N, et al. Front Microbiol. 2018 Oct 24;9:2470. doi: 10.3389/fmicb.2018.02470. eCollection 2018. Front Microbiol. 2018. PMID: 30405551 Free PMC article. - Petabase-scale sequence alignment catalyses viral discovery.
Edgar RC, Taylor B, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF, de la Peña M, Korobeynikov A, Chikhi R, Babaian A. Edgar RC, et al. Nature. 2022 Feb;602(7895):142-147. doi: 10.1038/s41586-021-04332-2. Epub 2022 Jan 26. Nature. 2022. PMID: 35082445
References
- Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
- Tringe SG, von Mering C, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources