Temporal dynamics and decay of putatively allochthonous and autochthonous viral genotypes in contrasting freshwater lakes - PubMed (original) (raw)
Temporal dynamics and decay of putatively allochthonous and autochthonous viral genotypes in contrasting freshwater lakes
Ian Hewson et al. Appl Environ Microbiol. 2012 Sep.
Abstract
Aquatic viruses play important roles in the biogeochemistry and ecology of lacustrine ecosystems; however, their composition, dynamics, and interactions with viruses of terrestrial origin are less extensively studied. We used a viral shotgun metagenomic approach to elucidate candidate autochthonous (i.e., produced within the lake) and allochthonous (i.e., washed in from other habitats) viral genotypes for a comparative study of their dynamics in lake waters. Based on shotgun metagenomes prepared from catchment soil and freshwater samples from two contrasting lakes (Cayuga Lake and Fayetteville Green Lake), we selected two putatively autochthonous viral genotypes (phycodnaviruses likely infecting algae and cyanomyoviruses likely infecting picocyanobacteria) and two putatively allochthonous viral genotypes (geminiviruses likely infecting terrestrial plants and circoviruses infecting unknown hosts but common in soil libraries) for analysis by genotype-specific quantitative PCR (TaqMan) applied to DNAs from viruses in the viral size fraction of lake plankton, i.e., 0.2 μm > virus > 0.02 μm. The abundance of autochthonous genotypes largely reflected expected host abundance, while the abundance of allochthonous genotypes corresponded with rainfall and storm events in the respective catchments, suggesting that viruses with these genotypes may have been transported to the lake in runoff. The decay rates of allochthonous and autochthonous genotypes, assessed in incubations where all potential hosts were killed, were generally lower (0.13 to 1.50% h(-1)) than those reported for marine virioplankton but similar to those for freshwater virioplankton. Both allochthonous and autochthonous viral genotypes were detected at higher concentrations in subsurface sediments than at the water-sediment interface. Our data indicate that putatively allochthonous viruses are present in lake plankton and sediments, where their temporal dynamics reflect active transport to the lake during hydrological events and then decay once there.
Figures
Fig 1
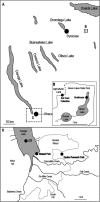
(A) Map of sampling locations in the Finger Lakes region of upstate New York. Samples were collected from two contrasting lakes: Fayetteville Green Lake, within the Green Lakes State Park, at the boathouse for plankton and sediments and on agricultural land nearby for soil (B); and Cayuga Lake, in Stewart Park and at site LC3 for plankton and at the equine research park for soil (C).
Fig 2
Phylogenetic annotation of metaviromes of sequence reads (A and B) and contiguous sequences (C). The read annotations were performed using the VIROME pipeline (
http://virome.diagcomputing.org/
), while the contig annotation was performed based on BLASTx searches of contig ORFs against viral, bacterial, archaeal, and eukaryotic databases at CAMERA (
), using an E value cutoff of 10−3. (A) Affiliation of reads by kingdom. (B) Viral read annotation by family. (C) Viral contig affiliation by family.
Fig 3
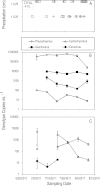
Catchment precipitation (A) and viral genotype abundances in Fayetteville Green Lake (FGL) (B) and Cayuga Lake (CL) (C) during summer 2011. Error bars show SE for duplicate samples. Autochthonous viral genotypes are indicated by open symbols, while allochthonous viral genotypes are indicated by closed symbols. Precipitation data were obtained for Ithaca and Syracuse weather stations from the National Climatic Data Service. Missing data at any time point indicate that abundances were below the detection threshold.
Fig 4
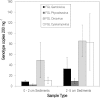
Abundance of viral genotypes in surface and deep sediments and within net plankton (>64 μm) in Fayetteville Green Lake. Error bars show SE for duplicate samples.
Similar articles
- Variations in abundance, genome size, morphology, and functional role of the virioplankton in Lakes Annecy and Bourget over a 1-year period.
Zhong X, Ram AS, Colombet J, Jacquet S. Zhong X, et al. Microb Ecol. 2014 Jan;67(1):66-82. doi: 10.1007/s00248-013-0320-2. Epub 2013 Nov 20. Microb Ecol. 2014. PMID: 24253662 - Nucleopolyhedrovirus detection and distribution in terrestrial, freshwater, and marine habitats of Appledore Island, Gulf of Maine.
Hewson I, Brown JM, Gitlin SA, Doud DF. Hewson I, et al. Microb Ecol. 2011 Jul;62(1):48-57. doi: 10.1007/s00248-011-9856-1. Epub 2011 Apr 21. Microb Ecol. 2011. PMID: 21509607 - Occurrence and seasonal dynamics of RNA viral genotypes in three contrasting temperate lakes.
Hewson I, Bistolas KSI, Button JB, Jackson EW. Hewson I, et al. PLoS One. 2018 Mar 15;13(3):e0194419. doi: 10.1371/journal.pone.0194419. eCollection 2018. PLoS One. 2018. PMID: 29543885 Free PMC article. - Humic substances-part 7: the biogeochemistry of dissolved organic carbon and its interactions with climate change.
Porcal P, Koprivnjak JF, Molot LA, Dillon PJ. Porcal P, et al. Environ Sci Pollut Res Int. 2009 Sep;16(6):714-26. doi: 10.1007/s11356-009-0176-7. Epub 2009 May 22. Environ Sci Pollut Res Int. 2009. PMID: 19462191 Review. - Viruses in Polar Lake and Soil Ecosystems.
Rastrojo A, Alcamí A. Rastrojo A, et al. Adv Virus Res. 2018;101:39-54. doi: 10.1016/bs.aivir.2018.02.002. Epub 2018 Apr 26. Adv Virus Res. 2018. PMID: 29908593 Review.
Cited by
- Genomic characteristics and environmental distributions of the uncultivated Far-T4 phages.
Roux S, Enault F, Ravet V, Pereira O, Sullivan MB. Roux S, et al. Front Microbiol. 2015 Mar 16;6:199. doi: 10.3389/fmicb.2015.00199. eCollection 2015. Front Microbiol. 2015. PMID: 25852662 Free PMC article. - Caught in the middle with multiple displacement amplification: the myth of pooling for avoiding multiple displacement amplification bias in a metagenome.
Marine R, McCarren C, Vorrasane V, Nasko D, Crowgey E, Polson SW, Wommack KE. Marine R, et al. Microbiome. 2014 Jan 30;2(1):3. doi: 10.1186/2049-2618-2-3. Microbiome. 2014. PMID: 24475755 Free PMC article. - Enumerating soil biodiversity.
Anthony MA, Bender SF, van der Heijden MGA. Anthony MA, et al. Proc Natl Acad Sci U S A. 2023 Aug 15;120(33):e2304663120. doi: 10.1073/pnas.2304663120. Epub 2023 Aug 7. Proc Natl Acad Sci U S A. 2023. PMID: 37549278 Free PMC article. Review. - Comparative study of viromes from freshwater samples of the Ile-Balkhash region of Kazakhstan captured through metagenomic analysis.
Alexyuk MS, Turmagambetova AS, Alexyuk PG, Bogoyavlenskiy AP, Berezin VE. Alexyuk MS, et al. Virusdisease. 2017 Mar;28(1):18-25. doi: 10.1007/s13337-016-0353-5. Epub 2017 Feb 1. Virusdisease. 2017. PMID: 28466051 Free PMC article. - Revealing viral diversity in the Napahai plateau wetland based on metagenomics.
Xiong L, Li Y, Zeng K, Wei Y, Li H, Ji X. Xiong L, et al. Antonie Van Leeuwenhoek. 2023 Dec 28;117(1):3. doi: 10.1007/s10482-023-01912-2. Antonie Van Leeuwenhoek. 2023. PMID: 38153618
References
- Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. 2005. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J. Aerosol Sci. 36:801–812
- Bergh O, Borsheim KY, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments. Nature 340:467–468 - PubMed
- Bettarel Y, Bouvier T, Bouvy M. 2009. Viral persistence in water as evaluated from a tropical/temperate cross-incubation. J. Plankton Res. 31:909–916
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources