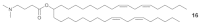Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo - PubMed (original) (raw)
. 2012 Aug 20;51(34):8529-33.
doi: 10.1002/anie.201203263. Epub 2012 Jul 10.
Steven M Ansell, Barbara L Mui, Ying K Tam, Jianxin Chen, Xinyao Du, David Butler, Laxman Eltepu, Shigeo Matsuda, Jayaprakash K Narayanannair, Kallanthottathil G Rajeev, Ismail M Hafez, Akin Akinc, Martin A Maier, Mark A Tracy, Pieter R Cullis, Thomas D Madden, Muthiah Manoharan, Michael J Hope
Affiliations
- PMID: 22782619
- PMCID: PMC3470698
- DOI: 10.1002/anie.201203263
Free PMC article
Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo
Muthusamy Jayaraman et al. Angew Chem Int Ed Engl. 2012.
Free PMC article
Abstract
Special (lipid) delivery: The role of the ionizable lipid pK(a) in the in vivo delivery of siRNA by lipid nanoparticles has been studied with a large number of head group modifications to the lipids. A tight correlation between the lipid pK(a) value and silencing of the mouse FVII gene (FVII ED(50) ) was found, with an optimal pK(a) range of 6.2-6.5. The most potent cationic lipid from this study has ED(50) levels around 0.005 mg kg(-1) in mice and less than 0.03 mg kg(-1) in non-human primates.
Copyright © 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
Figure 1
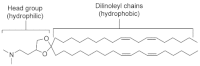
2,2-dilinoleyl-4-dimethylaminoethyl-[1,3]-dioxolane (DLin-KC2-DMA, 1). No difference in activity between enantiomers (4, 5) or the racemic mixture was observed (Table 1).
Figure 2
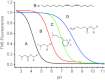
Determination of p_K_a by in situ TNS fluorescence titration. The four amino lipids shown are examples illustrating the full p_K_a range evaluated. Sigmoidal best-fit analyses were applied with p_K_a defined as the pH at half-maximal fluorescence intensity. A) 14, p_K_a=4.17; B) 52, p_K_a=5.73; C) 19, p_K_a=6.95; D) 48, p_K_a=8.12 (see Supporting Information page 40 for details).
Figure 3
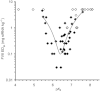
Plot of in vivo hepatic gene silencing activity vs. p_K_a in mice. The 56 amino lipids were formulated in LNPs and subjected to an ED50 analysis (described in Supporting Information, page 39) and plotted against their p_K_a. For 15 lipids the ED50 dose was not achieved, these are indicated by the open diamonds representing the highest dose tested for that lipid. For the remaining lipids (closed diamonds), a polynomial best-fit curve highlights the most active compounds, which exhibit an optimal p_K_a between 6.2 and 6.5. Each data point is derived from a four-dose response curve with groups of _n_=4 mice per dose.
Figure 4
(6_Z_,9_Z_,28_Z_,31_Z_)-Heptatriaconta-6,9,28,31-tetraen-19-yl 4-(dimethylamino)butanoate (16, Table 1).
Figure 5
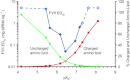
Plot of hepatic gene silencing activity in mice, p_K_a and calculated protonated/unprotonated amino lipid (% mole fractions) for the set of novel lipids between pKa 4 and 8.5. Each FVII ED50 data point (blue) is derived from a four-dose response (_n_=4 mice per dose level). Open diamonds represent lipids for which the ED50 was not achieved and are shown at the highest dose tested. The mole % of each amino lipid estimated to carry a positive charge in blood, assuming pH 7.4, is shown in red and the mole % amino lipid estimated to be uncharged in acidified endosomes at pH 5.5 is shown in green.
Figure 6
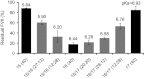
Lipid mixing supports p_K_a value as the dominant factor in determining in vivo hepatic gene-silencing activity for a given set of amino lipids. Mixtures of amino lipids 15, 16, and 17 were made to incrementally shift the average surface p_K_a value of the resulting LNP between 5.64 and 6.93. Numbers in parenthesis indicate the amount of each lipid in the mixture to make up the 40 mole % amino lipid component of the LNP. Bars represent group mean (_n_=4)±s.d.
Figure 7
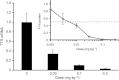
Efficacy of siRNA-LNP containing amino lipid 16 in mice and non-human primates. An optimized formulation composition containing 50 mole % of 16 was employed in this study (see text and Supporting Information page 41 for details). Inset: residual FVII activity in mouse serum relative to a saline control group with a median effective siRNA dose (ED50) of about 0.005 mg kg−1; data points represent group mean (_n_=5)±s.d. Cynomolgus monkeys were administered siRNA targeting the TTR gene at doses of 0.03, 0.1 and 0.3 mg kg−1 (bar graph). Data represent group mean (_n_=3)±s.d., expressed as TTR mRNA relative to GAPDH mRNA levels determined in liver samples, with the ED50 estimated to be <0.03 mg kg−1.
Similar articles
- Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells.
Basha G, Novobrantseva TI, Rosin N, Tam YY, Hafez IM, Wong MK, Sugo T, Ruda VM, Qin J, Klebanov B, Ciufolini M, Akinc A, Tam YK, Hope MJ, Cullis PR. Basha G, et al. Mol Ther. 2011 Dec;19(12):2186-200. doi: 10.1038/mt.2011.190. Epub 2011 Oct 4. Mol Ther. 2011. PMID: 21971424 Free PMC article. - Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA.
Chen S, Tam YYC, Lin PJC, Sung MMH, Tam YK, Cullis PR. Chen S, et al. J Control Release. 2016 Aug 10;235:236-244. doi: 10.1016/j.jconrel.2016.05.059. Epub 2016 May 26. J Control Release. 2016. PMID: 27238441 - Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates.
Suzuki Y, Hyodo K, Suzuki T, Tanaka Y, Kikuchi H, Ishihara H. Suzuki Y, et al. Int J Pharm. 2017 Mar 15;519(1-2):34-43. doi: 10.1016/j.ijpharm.2017.01.016. Epub 2017 Jan 9. Int J Pharm. 2017. PMID: 28089936 - Lipid nanoparticles for short interfering RNA delivery.
Leung AK, Tam YY, Cullis PR. Leung AK, et al. Adv Genet. 2014;88:71-110. doi: 10.1016/B978-0-12-800148-6.00004-3. Adv Genet. 2014. PMID: 25409604 Free PMC article. Review. - Lipid-based nanoparticles in the systemic delivery of siRNA.
Lin Q, Chen J, Zhang Z, Zheng G. Lin Q, et al. Nanomedicine (Lond). 2014 Jan;9(1):105-20. doi: 10.2217/nnm.13.192. Nanomedicine (Lond). 2014. PMID: 24354813 Review.
Cited by
- Nanomedicine-based strategies for the treatment of vein graft disease.
Zhou Z, Chen W, Cao Y, Abdi R, Tao W. Zhou Z, et al. Nat Rev Cardiol. 2024 Nov 5. doi: 10.1038/s41569-024-01094-y. Online ahead of print. Nat Rev Cardiol. 2024. PMID: 39501093 Review. - High-throughput synthesis and optimization of ionizable lipids through A3 coupling for efficient mRNA delivery.
Li J, Hu J, Jin D, Huo H, Chen N, Lin J, Lu X. Li J, et al. J Nanobiotechnology. 2024 Nov 4;22(1):672. doi: 10.1186/s12951-024-02919-1. J Nanobiotechnology. 2024. PMID: 39497197 Free PMC article. - Hydrophobic CPP/HDO conjugates: a new frontier in oligonucleotide-warheaded PROTAC delivery.
Naganuma M, Ohoka N, Hirano M, Watanabe D, Tsuji G, Inoue T, Demizu Y. Naganuma M, et al. RSC Med Chem. 2024 Sep 26. doi: 10.1039/d4md00546e. Online ahead of print. RSC Med Chem. 2024. PMID: 39421539 Free PMC article. - Engineering branched ionizable lipid for hepatic delivery of clustered regularly interspaced short palindromic repeat-Cas9 ribonucleoproteins.
Onuma H, Shimizu R, Suzuki Y, Sato M, Harashima H, Sato Y. Onuma H, et al. iScience. 2024 Sep 11;27(10):110928. doi: 10.1016/j.isci.2024.110928. eCollection 2024 Oct 18. iScience. 2024. PMID: 39381750 Free PMC article. - Ionizable Cationic Lipids and Helper Lipids Synergistically Contribute to RNA Packing and Protection in Lipid-Based Nanomaterials.
Zimmer DN, Schmid F, Settanni G. Zimmer DN, et al. J Phys Chem B. 2024 Oct 17;128(41):10165-10177. doi: 10.1021/acs.jpcb.4c05057. Epub 2024 Oct 4. J Phys Chem B. 2024. PMID: 39366669
References
- Zimmermann TS, et al. Nature. 2006;441:111–114. - PubMed
- Semple SC, et al. Nat. Biotechnol. 2010;28:172–176. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources