Autophagy in the regulation of pathogen replication and adaptive immunity - PubMed (original) (raw)
Review
Autophagy in the regulation of pathogen replication and adaptive immunity
Felix Randow et al. Trends Immunol. 2012 Oct.
Abstract
Autophagy is an evolutionarily conserved homeostatic process by which cells deliver cytoplasmic material for degradation into lysosomes. Autophagy may have evolved as a nutrient-providing homeostatic pathway induced upon starvation, but with the acquisition of cargo receptors, autophagy has become an important cellular defence mechanism as well as a generator of antigenic peptides for major histocompatibility complex (MHC) presentation. We propose that autophagy efficiently protects against microbes encountering the cytosolic environment accidentally, for example, upon phagosomal damage, whereas pathogens routinely accessing the host cytosol have evolved to avoid or even benefit from autophagy.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
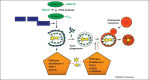
Figure 1
Restriction of pathogens by macroautophagy and their escape. Autophagosomes are formed with the help of two ubiquitin-like systems (ATG8/LC3 and ATG12) and their formation and degradation is guided by phosphatidylinositol-3 (PI3) kinase complexes. The ATG8/LC3 ligase complex ATG5–ATG12–ATG16L1 conjugates ATG8/LC3 to the isolation membrane or phagophore and is then recycled from the outer autophagosomal membrane with ATG8/LC3. Upon fusion with lysosomes, the inner autophagosomal membrane and its content, including pathogens, is degraded. Viruses interfere with this process and either block autophagosome formation or degradation by interacting with ATG6/BECLIN-1, which is contained in autophagosome forming PI3 kinase complexes (VPS34 as PI3 kinase, VPS15, ATG6/BECLIN-1 and ATG14L) and autophagosome degrading (VPS34, VPS15, ATG6/BECLIN-1 and UVRAG) or degradation blocking (VPS34, VPS15, ATG6/BECLIN-1, UVRAG and Rubicon) PI3 kinase complexes.
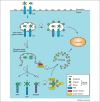
Figure 2
How autophagy and LC3-assisted phagocytosis defend cells against infection. Synergistic lines of defence prevent the entry of pathogens into the cytosol of host cells. (a) LC3-assisted phagocytosis is triggered by Toll-like receptors (TLRs) and potentially other pattern recognition receptors (PRRs) in response to microorganisms that are taken up by phagocytosis or that have actively invaded nonphagocytic cells. LC3-assisted phagocytosis requires a subset of autophagy genes for the labelling of phagosomes with ATG8/LC3, which promotes their lysosomal delivery and the efficient killing of vesicular pathogens. (b) Damage to the limiting membrane of the pathogen-containing vesicle, either accidental or caused by pathogens attempting to escape from the vesicle, exposes the cytosol to glycans previously hidden inside the vesicle. (c) Cytosol-accessible glycans are detected by the danger receptor galectin-8, which, by recruiting the cargo receptor NDP52 (nuclear dot protein 52), triggers autophagy. (d) Pathogens having escaped galectin-8-induced autophagy are met by yet another layer of PRRs in the cytosol. (e) A yet-to-be-identified E3 ubiquitin ligase causes the ubiquitin-coating of invading bacteria. It remains to be established whether this ligase only targets membrane-associated or also free-floating bacteria, whether it is a PRR, and also whether its substrate is of bacterial or host origin. (f) Ubiquitin-coated bacteria are targeted for autophagy by three apparently non-redundant cargo receptors, that is, NDP52, p62, and optineurin.
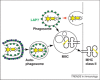
Figure 3
Antigen for major histocompatibility complex (MHC) class II presentation can be provided by phagocytosis or macroautophagy. Cytosolic pathogens are delivered to MHC class II containing compartments (MIICs) via macroautophagy for antigen loading. Extracellular pathogens reach this MHC class II loading compartment via endocytosis, which might be in some instances be facilitated by ATG8/LC3 coupled to the phagosome.
Similar articles
- Antigen Presentation and Autophagy in Teleost Adaptive Immunity.
Johnstone C, Chaves-Pozo E. Johnstone C, et al. Int J Mol Sci. 2022 Apr 28;23(9):4899. doi: 10.3390/ijms23094899. Int J Mol Sci. 2022. PMID: 35563287 Free PMC article. Review. - Manipulation of Autophagy by Bacterial Pathogens Impacts Host Immunity.
Kunz TC, Viana F, Buchrieser C, Escoll P. Kunz TC, et al. Curr Issues Mol Biol. 2018;25:81-98. doi: 10.21775/cimb.025.081. Epub 2017 Sep 6. Curr Issues Mol Biol. 2018. PMID: 28875941 Review. - Autophagy in CD4+ T-cell immunity and tolerance.
Lünemann JD, Münz C. Lünemann JD, et al. Cell Death Differ. 2009 Jan;16(1):79-86. doi: 10.1038/cdd.2008.113. Epub 2008 Jul 18. Cell Death Differ. 2009. PMID: 18636073 Review. - Autophagy in immunity and cell-autonomous defense against intracellular microbes.
Deretic V. Deretic V. Immunol Rev. 2011 Mar;240(1):92-104. doi: 10.1111/j.1600-065X.2010.00995.x. Immunol Rev. 2011. PMID: 21349088 Free PMC article. Review. - Autophagy in innate and adaptive immunity against intracellular pathogens.
Schmid D, Dengjel J, Schoor O, Stevanovic S, Münz C. Schmid D, et al. J Mol Med (Berl). 2006 Mar;84(3):194-202. doi: 10.1007/s00109-005-0014-4. Epub 2006 Jan 28. J Mol Med (Berl). 2006. PMID: 16501849 Review.
Cited by
- The infection characteristics and autophagy defect of dermal macrophages in STZ-induced diabetic rats skin wound Staphylococcus aureus infection model.
Xie X, Zhong R, Luo L, Lin X, Huang L, Huang S, Ni L, Chen B, Shen R, Yan L, Duan C. Xie X, et al. Immun Inflamm Dis. 2021 Dec;9(4):1428-1438. doi: 10.1002/iid3.492. Epub 2021 Oct 14. Immun Inflamm Dis. 2021. PMID: 34647429 Free PMC article. - Historical landmarks of autophagy research.
Ohsumi Y. Ohsumi Y. Cell Res. 2014 Jan;24(1):9-23. doi: 10.1038/cr.2013.169. Epub 2013 Dec 24. Cell Res. 2014. PMID: 24366340 Free PMC article. Review. - Insights into Autophagy in Microbiome Therapeutic Approaches for Drug-Resistant Tuberculosis.
Rahim MA, Seo H, Barman I, Hossain MS, Shuvo MSH, Song HY. Rahim MA, et al. Cells. 2025 Apr 3;14(7):540. doi: 10.3390/cells14070540. Cells. 2025. PMID: 40214493 Free PMC article. Review. - TLR and NLRP3 inflammasome-dependent innate immune responses to tumor-derived autophagosomes (DRibbles).
Xing Y, Cao R, Hu HM. Xing Y, et al. Cell Death Dis. 2016 Aug 4;7(8):e2322. doi: 10.1038/cddis.2016.206. Cell Death Dis. 2016. PMID: 27490927 Free PMC article. - Innate immunity to adenovirus.
Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A, Greber UF. Hendrickx R, et al. Hum Gene Ther. 2014 Apr;25(4):265-84. doi: 10.1089/hum.2014.001. Epub 2014 Apr 8. Hum Gene Ther. 2014. PMID: 24512150 Free PMC article. Review.
References
- Mizushima N. The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. - PubMed
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_U105170648/MRC_/Medical Research Council/United Kingdom
- R01 CA108609/CA/NCI NIH HHS/United States
- R01CA108609/CA/NCI NIH HHS/United States
- U10517064/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials