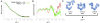Integrative structural modeling with small angle X-ray scattering profiles - PubMed (original) (raw)
Review
Integrative structural modeling with small angle X-ray scattering profiles
Dina Schneidman-Duhovny et al. BMC Struct Biol. 2012.
Abstract
Recent technological advances enabled high-throughput collection of Small Angle X-ray Scattering (SAXS) profiles of biological macromolecules. Thus, computational methods for integrating SAXS profiles into structural modeling are needed more than ever. Here, we review specifically the use of SAXS profiles for the structural modeling of proteins, nucleic acids, and their complexes. First, the approaches for computing theoretical SAXS profiles from structures are presented. Second, computational methods for predicting protein structures, dynamics of proteins in solution, and assembly structures are covered. Third, we discuss the use of SAXS profiles in integrative structure modeling approaches that depend simultaneously on several data types.
Figures
Figure 1
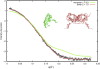
Theoretical SAXS profiles of glucose isomerase from five programs fitted against the experimental SAXS profile.
Figure 2
Yeast Nup145N (443–605) crystal structure [PDB:3kep] monomer (green) and dimer (red) versus the solution SAXS profile (black).
Figure 3
FoXS-MES server output for ensemble fit (green) of the SAXS profile (black) versus a single conformation fit (red) for XLF-XRCC4 filaments. A) Fit plot, B) Residuals plot, and C) Ensemble structures and weights.
Figure 4
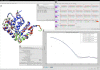
Comparative modeling and SAXS profile fitting using the Chimera visualization package. MODELLER was used to add missing residues and a His tag on the C-terminal domain of Nup133 (see sequence alignment in the upper right window). Three models were generated (red, green and blue) using a template structure [PDB:3KFO] (white). The FoXS interface (lower left window) was used to fit the profiles computed for the X-ray structure and the models to the experimental SAXS profile (lower right window). A) model window, B) sequence alignment window, C) model scores, D) FoXS interface window, and E) FoXS output window.
Figure 5
FoXS-MES ensemble fit (green) of the SAXS profile (black) versus a single conformation fit (red) for Mre11-Rad50. A) Fit plot, B) Residuals plot, and C) Ensemble structures and weights.
Figure 6
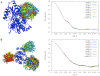
Docking models with similar shapes generate similar profiles. A) Top 10 docking models for adrenodoxin reductase-adrenodoxin complex [PDB:1E6E], and B) top 20 docking models for PAPS reductase-thioredoxin complex [PDB:2O8V].
Similar articles
- Hybrid Methods for Modeling Protein Structures Using Molecular Dynamics Simulations and Small-Angle X-Ray Scattering Data.
Ekimoto T, Ikeguchi M. Ekimoto T, et al. Adv Exp Med Biol. 2018;1105:237-258. doi: 10.1007/978-981-13-2200-6_15. Adv Exp Med Biol. 2018. PMID: 30617833 Review. - Multi-state modeling of antibody-antigen complexes with SAXS profiles and deep-learning models.
Cohen T, Halfon M, Carter L, Sharkey B, Jain T, Sivasubramanian A, Schneidman-Duhovny D. Cohen T, et al. Methods Enzymol. 2023;678:237-262. doi: 10.1016/bs.mie.2022.11.003. Epub 2022 Dec 8. Methods Enzymol. 2023. PMID: 36641210 - Modeling Structure and Dynamics of Protein Complexes with SAXS Profiles.
Schneidman-Duhovny D, Hammel M. Schneidman-Duhovny D, et al. Methods Mol Biol. 2018;1764:449-473. doi: 10.1007/978-1-4939-7759-8_29. Methods Mol Biol. 2018. PMID: 29605933 Free PMC article. - Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies.
Förster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Förster F, et al. J Mol Biol. 2008 Oct 17;382(4):1089-106. doi: 10.1016/j.jmb.2008.07.074. Epub 2008 Jul 31. J Mol Biol. 2008. PMID: 18694757 Free PMC article. - Developments in small-angle X-ray scattering (SAXS) for characterizing the structure of surfactant-macromolecule interactions and their complex.
Chen R, Song Y, Wang Z, Ji H, Du Z, Ma Q, Yang Y, Liu X, Li N, Sun Y. Chen R, et al. Int J Biol Macromol. 2023 Nov 1;251:126288. doi: 10.1016/j.ijbiomac.2023.126288. Epub 2023 Aug 13. Int J Biol Macromol. 2023. PMID: 37582436 Review.
Cited by
- Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data.
Aprahamian ML, Chea EE, Jones LM, Lindert S. Aprahamian ML, et al. Anal Chem. 2018 Jun 19;90(12):7721-7729. doi: 10.1021/acs.analchem.8b01624. Epub 2018 Jun 6. Anal Chem. 2018. PMID: 29874044 Free PMC article. - Disentangling polydispersity in the PCNA-p15PAF complex, a disordered, transient and multivalent macromolecular assembly.
Cordeiro TN, Chen PC, De Biasio A, Sibille N, Blanco FJ, Hub JS, Crehuet R, Bernadó P. Cordeiro TN, et al. Nucleic Acids Res. 2017 Feb 17;45(3):1501-1515. doi: 10.1093/nar/gkw1183. Nucleic Acids Res. 2017. PMID: 28180305 Free PMC article. - Universally Accessible Structural Data on Macromolecular Conformation, Assembly, and Dynamics by Small Angle X-Ray Scattering for DNA Repair Insights.
Chinnam NB, Syed A, Burnett KH, Hura GL, Tainer JA, Tsutakawa SE. Chinnam NB, et al. Methods Mol Biol. 2022;2444:43-68. doi: 10.1007/978-1-0716-2063-2_4. Methods Mol Biol. 2022. PMID: 35290631 Free PMC article. - Combining small angle X-ray scattering (SAXS) with protein structure predictions to characterize conformations in solution.
Chinnam NB, Syed A, Hura GL, Hammel M, Tainer JA, Tsutakawa SE. Chinnam NB, et al. Methods Enzymol. 2023;678:351-376. doi: 10.1016/bs.mie.2022.09.023. Epub 2022 Oct 31. Methods Enzymol. 2023. PMID: 36641214 Free PMC article. - Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy.
Ogorzalek TL, Hura GL, Belsom A, Burnett KH, Kryshtafovych A, Tainer JA, Rappsilber J, Tsutakawa SE, Fidelis K. Ogorzalek TL, et al. Proteins. 2018 Mar;86 Suppl 1(Suppl 1):202-214. doi: 10.1002/prot.25452. Epub 2018 Feb 7. Proteins. 2018. PMID: 29314274 Free PMC article.
References
- Svergun DI. Small-angle X-ray and neutron scattering as a tool for structural systems biology. Biol Chem. 2010;391(7):737–743. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources