Reconstructing Native American population history - PubMed (original) (raw)
. 2012 Aug 16;488(7411):370-4.
doi: 10.1038/nature11258.
Nick Patterson, Desmond Campbell, Arti Tandon, Stéphane Mazieres, Nicolas Ray, Maria V Parra, Winston Rojas, Constanza Duque, Natalia Mesa, Luis F García, Omar Triana, Silvia Blair, Amanda Maestre, Juan C Dib, Claudio M Bravi, Graciela Bailliet, Daniel Corach, Tábita Hünemeier, Maria Cátira Bortolini, Francisco M Salzano, María Luiza Petzl-Erler, Victor Acuña-Alonzo, Carlos Aguilar-Salinas, Samuel Canizales-Quinteros, Teresa Tusié-Luna, Laura Riba, Maricela Rodríguez-Cruz, Mardia Lopez-Alarcón, Ramón Coral-Vazquez, Thelma Canto-Cetina, Irma Silva-Zolezzi, Juan Carlos Fernandez-Lopez, Alejandra V Contreras, Gerardo Jimenez-Sanchez, Maria José Gómez-Vázquez, Julio Molina, Angel Carracedo, Antonio Salas, Carla Gallo, Giovanni Poletti, David B Witonsky, Gorka Alkorta-Aranburu, Rem I Sukernik, Ludmila Osipova, Sardana A Fedorova, René Vasquez, Mercedes Villena, Claudia Moreau, Ramiro Barrantes, David Pauls, Laurent Excoffier, Gabriel Bedoya, Francisco Rothhammer, Jean-Michel Dugoujon, Georges Larrouy, William Klitz, Damian Labuda, Judith Kidd, Kenneth Kidd, Anna Di Rienzo, Nelson B Freimer, Alkes L Price, Andrés Ruiz-Linares
Affiliations
- PMID: 22801491
- PMCID: PMC3615710
- DOI: 10.1038/nature11258
Reconstructing Native American population history
David Reich et al. Nature. 2012.
Erratum in
- Nature. 2012 Nov 8;491(7423):288
Abstract
The peopling of the Americas has been the subject of extensive genetic, archaeological and linguistic research; however, central questions remain unresolved. One contentious issue is whether the settlement occurred by means of a single migration or multiple streams of migration from Siberia. The pattern of dispersals within the Americas is also poorly understood. To address these questions at a higher resolution than was previously possible, we assembled data from 52 Native American and 17 Siberian groups genotyped at 364,470 single nucleotide polymorphisms. Here we show that Native Americans descend from at least three streams of Asian gene flow. Most descend entirely from a single ancestral population that we call 'First American'. However, speakers of Eskimo-Aleut languages from the Arctic inherit almost half their ancestry from a second stream of Asian gene flow, and the Na-Dene-speaking Chipewyan from Canada inherit roughly one-tenth of their ancestry from a third stream. We show that the initial peopling followed a southward expansion facilitated by the coast, with sequential population splits and little gene flow after divergence, especially in South America. A major exception is in Chibchan speakers on both sides of the Panama isthmus, who have ancestry from both North and South America.
Figures
Figure 1. Geographic, linguistic and genetic overview of 52 Native American populations
(A) Sampling locations of the populations, with colors corresponding to linguistic groups. (B) Cluster-based analysis (k=4) using ADMIXTURE shows evidence of some West Eurasian-related and sub-Saharan-African-related ancestry in many Native Americans prior to masking (top), but little afterward (bottom). Thick vertical lines denote major linguistic groupings, and thin vertical lines separate individual populations. (C) Neighbor-Joining tree based on FST distances relating Native American to selected non-American populations (sample sizes in parentheses). Native American and Siberian data were analyzed after masking but consistent trees were obtained on a subset of completely unadmixed samples (Figure S3). Some populations have evidence for substructure, and we represent these as two different groups (e.g. Maya1 and Maya2).
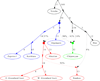
Figure 2. Distinct streams of gene flow from Asia into America
We present an Admixture Graph (AG) that gives no evidence of being a poor fit to the data and is consistent with three streams of Asian gene flow into America. Solid points indicate inferred ancestral populations; drift on each lineage is given in units proportional to 1000×FST; and mixture events (dotted lines) are denoted by the percentage of ancestry. The Asian lineage leading to First Americans is the most deeply diverged, while the Asian lineages leading to Eskimo-Aleut speakers and the Na-Dene speaking Chipewyan are more closely related and descend from a common Siberian ancestral population that is a sister group to the Han. The inferred ancestral populations are indicated by filled circles and the lineages descending from them are colored: First American (blue), ancestors of the Na-Dene speaking Chipewyan (green) and Eskimo-Aleut (red). The model also infers a migration of people related to Eskimo-Aleut speakers across the Bering Strait, thus bringing First American genes to Asia (the Naukan are shown, but the Chukchi show a similar pattern; Note S7). Estimated admixture proportions are shown along the dotted lines, and lineage-specific drift estimates are in units proportional to 1000×FST
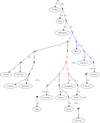
Figure 3. A model fitting populations of entirely First American ancestry
We show an Admixture Graph (AG) depicting the relationships among 16 selected Native American populations with entirely First American ancestry along with 2 outgroups (Yoruba and Han). The Colombian Inga are modeled as a mixture of Andean and Amazonian ancestry. The Paraguayan Guarani are fit as a mixture of separate strands of ancestry from eastern South America. The Central American Cabecar are modeled as a mixture of strands of ancestry related to South Americans and to North Americans, supporting back-migration from South into Central America. The coloring of edges indicates alternative insertion points for the admixing lineages leading to the Cabecar that produce a similar fit to the data in the sense that the χ2 statistic is within 3.84 of the AG shown. The red coloring shows that the South American lineage contributing to the Cabecar split off after the divergence of the Andean populations, and the blue coloring shows that the other lineage present in the Cabecar diverged before the separation of Andeans.
Similar articles
- Y-chromosome evidence for differing ancient demographic histories in the Americas.
Bortolini MC, Salzano FM, Thomas MG, Stuart S, Nasanen SP, Bau CH, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-de-Guerra D, Torres MM, Groot H, Michalski R, Nymadawa P, Bedoya G, Bradman N, Labuda D, Ruiz-Linares A. Bortolini MC, et al. Am J Hum Genet. 2003 Sep;73(3):524-39. doi: 10.1086/377588. Epub 2003 Jul 28. Am J Hum Genet. 2003. PMID: 12900798 Free PMC article. - Late Pleistocene exploration and settlement of the Americas by modern humans.
Waters MR. Waters MR. Science. 2019 Jul 12;365(6449):eaat5447. doi: 10.1126/science.aat5447. Science. 2019. PMID: 31296740 Review. - Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: evolutionary and demographic evidence.
Schurr TG, Sherry ST. Schurr TG, et al. Am J Hum Biol. 2004 Jul-Aug;16(4):420-39. doi: 10.1002/ajhb.20041. Am J Hum Biol. 2004. PMID: 15214060 - A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data.
Bonatto SL, Salzano FM. Bonatto SL, et al. Proc Natl Acad Sci U S A. 1997 Mar 4;94(5):1866-71. doi: 10.1073/pnas.94.5.1866. Proc Natl Acad Sci U S A. 1997. PMID: 9050871 Free PMC article. - The late Pleistocene dispersal of modern humans in the Americas.
Goebel T, Waters MR, O'Rourke DH. Goebel T, et al. Science. 2008 Mar 14;319(5869):1497-502. doi: 10.1126/science.1153569. Science. 2008. PMID: 18339930 Review.
Cited by
- Hemochromatosis neural archetype reveals iron disruption in motor circuits.
Loughnan R, Ahern J, Boyle M, Jernigan TL, Hagler DJ Jr, Iversen JR, Frei O, Smith DM, Andreassen O, Zaitlen N, Sugrue L, Thompson WK, Dale A, Schork AJ, Fan CC. Loughnan R, et al. Sci Adv. 2024 Nov 22;10(47):eadp4431. doi: 10.1126/sciadv.adp4431. Epub 2024 Nov 22. Sci Adv. 2024. PMID: 39576859 Free PMC article. - AN ANCIENT DNA PACIFIC JOURNEY: A CASE STUDY OF COLLABORATION BETWEEN ARCHAEOLOGISTS AND GENETICISTS.
Spriggs M, Reich D. Spriggs M, et al. World Archaeol. 2019;51(4):620-639. doi: 10.1080/00438243.2019.1733069. Epub 2020 Mar 17. World Archaeol. 2019. PMID: 39564545 Free PMC article. - Unveiling ancestral threads: Exploring CCR5 ∆32 mutation frequencies in Colombian populations for HIV/AIDS therapeutics.
Barrios-Navas A, Nguyen TL, Gallo JE, Mariño-Ramírez L, Soto JMS, Sánchez A, Jordan IK, Valderrama-Aguirre A. Barrios-Navas A, et al. Infect Genet Evol. 2024 Nov;125:105680. doi: 10.1016/j.meegid.2024.105680. Epub 2024 Oct 5. Infect Genet Evol. 2024. PMID: 39374819 - 9,000 years of genetic continuity in southernmost Africa demonstrated at Oakhurst rockshelter.
Gretzinger J, Gibbon VE, Penske SE, Sealy JC, Rohrlach AB, Salazar-García DC, Krause J, Schiffels S. Gretzinger J, et al. Nat Ecol Evol. 2024 Nov;8(11):2121-2134. doi: 10.1038/s41559-024-02532-3. Epub 2024 Sep 19. Nat Ecol Evol. 2024. PMID: 39300260 Free PMC article. - Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas.
Moreno-Mayar JV, Sousa da Mota B, Higham T, Klemm S, Gorman Edmunds M, Stenderup J, Iraeta-Orbegozo M, Laborde V, Heyer E, Torres Hochstetter F, Friess M, Allentoft ME, Schroeder H, Delaneau O, Malaspinas AS. Moreno-Mayar JV, et al. Nature. 2024 Sep;633(8029):389-397. doi: 10.1038/s41586-024-07881-4. Epub 2024 Sep 11. Nature. 2024. PMID: 39261618 Free PMC article.
References
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, UP: 1994.
- Meltzer DJ. First peoples in a new world : colonizing ice age America. University of California Press; 2009.
- Goebel T, Waters MR, O'Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319:1497–1502. - PubMed
- O'Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr. Biol. 2010;20:R202–R207. - PubMed
Publication types
MeSH terms
Grants and funding
- GM079558/GM/NIGMS NIH HHS/United States
- CAPMC/ CIHR/Canada
- R01 HG006399/HG/NHGRI NIH HHS/United States
- GM079558-S1/GM/NIGMS NIH HHS/United States
- BB/1021213/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- NS043538/NS/NINDS NIH HHS/United States
- R21 DK073818/DK/NIDDK NIH HHS/United States
- R01 MH075007/MH/NIMH NIH HHS/United States
- P01 GM057672/GM/NIGMS NIH HHS/United States
- R01 NS037484/NS/NINDS NIH HHS/United States
- R01 NS043538/NS/NINDS NIH HHS/United States
- R01 GM100233/GM/NIGMS NIH HHS/United States
- NS037484/NS/NINDS NIH HHS/United States
- HG006399/HG/NHGRI NIH HHS/United States
- MH075007/MH/NIMH NIH HHS/United States
- R01 GM079558/GM/NIGMS NIH HHS/United States
- GM057672/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous