Cryptic species in putative ancient asexual darwinulids (Crustacea, Ostracoda) - PubMed (original) (raw)
Cryptic species in putative ancient asexual darwinulids (Crustacea, Ostracoda)
Isa Schön et al. PLoS One. 2012.
Abstract
Background: Fully asexually reproducing taxa lack outcrossing. Hence, the classic Biological Species Concept cannot be applied.
Methodology/principal findings: We used DNA sequences from the mitochondrial COI gene and the nuclear ITS2 region to check species boundaries according to the evolutionary genetic (EG) species concept in five morphospecies in the putative ancient asexual ostracod genera, Penthesilenula and Darwinula, from different continents. We applied two methods for detecting cryptic species, namely the K/θ method and the General Mixed Yule Coalescent model (GMYC). We could confirm the existence of species in all five darwinulid morphospecies and additional cryptic diversity in three morphospecies, namely in Penthesilenula brasiliensis, Darwinula stevensoni and in P. aotearoa. The number of cryptic species within one morphospecies varied between seven (P. brasiliensis), five to six (D. stevensoni) and two (P. aotearoa), respectively, depending on the method used. Cryptic species mainly followed continental distributions. We also found evidence for coexistence at the local scale for Brazilian cryptic species of P. brasiliensis and P. aotearoa. Our ITS2 data confirmed that species exist in darwinulids but detected far less EG species, namely two to three cryptic species in P. brasiliensis and no cryptic species at all in the other darwinulid morphospecies.
Conclusions/significance: Our results clearly demonstrate that both species and cryptic diversity can be recognized in putative ancient asexual ostracods using the EG species concept, and that COI data are more suitable than ITS2 for this purpose. The discovery of up to eight cryptic species within a single morphospecies will significantly increase estimates of biodiversity in this asexual ostracod group. Which factors, other than long-term geographic isolation, are important for speciation processes in these ancient asexuals remains to be investigated.
Conflict of interest statement
Competing Interests: SH is employed by Bennelongia. This does not alter the authors’ adherence to all the PLoS ONE policies on sharing data and materials.
Figures
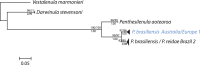
Figure 1. COI phylogeny of the three darwinulid genera.
This phylogeny was obtained by Bayesian Inference. Height of triangles reflects the number of sequences. Triangles or single branches represent EG species, which are indicated by their geographic origin and Arabic numbers. Species names in red are split into two species by the GMYC method (but not by the K/θ method). Species names in black are recognized by both methods. Numbers above branches are bootstrap values of 1000 replicates for NJ and ML trees (>50%), numbers below branches are posterior probabilities of Bayesian Inference.
Figure 2. ITS phylogeny of the three darwinulid genera.
This phylogeny was obtained by Bayesian Inference. Height of triangles reflects the number of sequences. Triangles or single branches represent EG species, which are indicated by their geographic origin and Arabic numbers. Species names in blue are only recognized by the K/θ method (but not by the GMYC method). Species names in black are recognized by both methods. Numbers above branches are bootstrap values of 1000 replicates for NJ and ML trees (>50%), numbers below branches are posterior probabilities of Bayesian Inference.
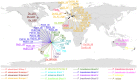
Figure 3. Global distribution of the EG species, determined by COI sequence data.
Letters and numbers in the map refer to the analyzed specimens (see Table S1 for more details). Different EG species are indicated by different color codes.
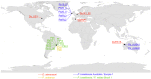
Figure 4. Global distribution of the EG species, determined by ITS2 sequence data.
Letters and numbers in the map refer to the analyzed specimens (see Table S2 for more details). Different EG species are indicated by different color codes.
Similar articles
- No slave to sex.
Schön I, Martens K. Schön I, et al. Proc Biol Sci. 2003 Apr 22;270(1517):827-33. doi: 10.1098/rspb.2002.2314. Proc Biol Sci. 2003. PMID: 12737661 Free PMC article. - Exceptional cryptic diversity and multiple origins of parthenogenesis in a freshwater ostracod.
Bode SN, Adolfsson S, Lamatsch DK, Martins MJ, Schmit O, Vandekerkhove J, Mezquita F, Namiotko T, Rossetti G, Schön I, Butlin RK, Martens K. Bode SN, et al. Mol Phylogenet Evol. 2010 Feb;54(2):542-52. doi: 10.1016/j.ympev.2009.08.022. Epub 2009 Aug 22. Mol Phylogenet Evol. 2010. PMID: 19703573 - Clonal diversity in the ancient asexual ostracod Darwinula stevensoni assessed by RAPD-PCR.
Van Doninck K, Schön I, Martens K, Backeljau T. Van Doninck K, et al. Heredity (Edinb). 2004 Aug;93(2):154-60. doi: 10.1038/sj.hdy.6800486. Heredity (Edinb). 2004. PMID: 15241465 - Ostracod (Ostracoda, Crustacea) genomics - Promises and challenges.
Schön I, Martens K. Schön I, et al. Mar Genomics. 2016 Oct;29:19-25. doi: 10.1016/j.margen.2016.03.008. Epub 2016 Mar 25. Mar Genomics. 2016. PMID: 27020380 Review. - Epigenetic variation in asexually reproducing organisms.
Verhoeven KJ, Preite V. Verhoeven KJ, et al. Evolution. 2014 Mar;68(3):644-55. doi: 10.1111/evo.12320. Epub 2013 Dec 13. Evolution. 2014. PMID: 24274255 Review.
Cited by
- Barcoding of ancient lake ostracods (crustacea) reveals cryptic speciation with extremely low distances.
Karanovic I. Karanovic I. PLoS One. 2015 Mar 26;10(3):e0121133. doi: 10.1371/journal.pone.0121133. eCollection 2015. PLoS One. 2015. PMID: 25811597 Free PMC article. - First annotated draft genomes of nonmarine ostracods (Ostracoda, Crustacea) with different reproductive modes.
Tran Van P, Anselmetti Y, Bast J, Dumas Z, Galtier N, Jaron KS, Martens K, Parker DJ, Robinson-Rechavi M, Schwander T, Simion P, Schön I. Tran Van P, et al. G3 (Bethesda). 2021 Apr 15;11(4):jkab043. doi: 10.1093/g3journal/jkab043. G3 (Bethesda). 2021. PMID: 33591306 Free PMC article. - Convergent evolution of aquatic life by sexual and parthenogenetic oribatid mites.
Krause A, Pachl P, Schulz G, Lehmitz R, Seniczak A, Schaefer I, Scheu S, Maraun M. Krause A, et al. Exp Appl Acarol. 2016 Dec;70(4):439-453. doi: 10.1007/s10493-016-0089-3. Epub 2016 Oct 26. Exp Appl Acarol. 2016. PMID: 27785647 - "Jack-of-all-trades" is parthenogenetic.
Maraun M, Bischof PSP, Klemp FL, Pollack J, Raab L, Schmerbach J, Schaefer I, Scheu S, Caruso T. Maraun M, et al. Ecol Evol. 2022 Jun 23;12(6):e9036. doi: 10.1002/ece3.9036. eCollection 2022 Jul. Ecol Evol. 2022. PMID: 35784052 Free PMC article. - From morphology to molecules: a combined source approach to untangle the taxonomy of Clessinia (Gastropoda, Odontostomidae), endemic land snails from the Dry Chaco ecoregion.
Cuezzo MG, Miranda MJ, Vogler RE, Beltramino AA. Cuezzo MG, et al. PeerJ. 2018 Dec 6;6:e5986. doi: 10.7717/peerj.5986. eCollection 2018. PeerJ. 2018. PMID: 30564514 Free PMC article.
References
- De Queiroz K. Species concepts and delimitation. System Biol. 2007;56:879–886. - PubMed
- Foottit RG. Claridge MF, Dawah H, Wilson MR, editors. Recognition of parthenogenetic insect species. 1997. pp. 291–308. Species, the units of biodiversity. Oxford: Chapman and Hall.
- Martens K, Rossetti G, Baltanas A. Martens K, editor. Reproductive modes and taxonomy. 1998. pp. 197–214. editor. Sex and Parthenogenesis – evolutionary ecology of reproductive modes in non-marine ostracods. Leiden: Backhuys Publishers.
- Loxdale H, Schön I, Martens K. Schön I, Martens K, Van Dijk P, editors. The elusive clone. 2009. pp. 187–200. Lost sex. The evolutionary biology of parthenogenesis. Dordrecht: Springer Academic Publishers.
- Mayr E. Populations, species and evolution. Harvard: Harvard University Press. 1970.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources