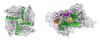Detecting pore-lining regions in transmembrane protein sequences - PubMed (original) (raw)
Detecting pore-lining regions in transmembrane protein sequences
Timothy Nugent et al. BMC Bioinformatics. 2012.
Abstract
Background: Alpha-helical transmembrane channel and transporter proteins play vital roles in a diverse range of essential biological processes and are crucial in facilitating the passage of ions and molecules across the lipid bilayer. However, the experimental difficulties associated with obtaining high quality crystals has led to their significant under-representation in structural databases. Computational methods that can identify structural features from sequence alone are therefore of high importance.
Results: We present a method capable of automatically identifying pore-lining regions in transmembrane proteins from sequence information alone, which can then be used to determine the pore stoichiometry. By labelling pore-lining residues in crystal structures using geometric criteria, we have trained a support vector machine classifier to predict the likelihood of a transmembrane helix being involved in pore formation. Results from testing this approach under stringent cross-validation indicate that prediction accuracy of 72% is possible, while a support vector regression model is able to predict the number of subunits participating in the pore with 62% accuracy.
Conclusion: To our knowledge, this is the first tool capable of identifying pore-lining regions in proteins and we present the results of applying it to a data set of sequences with available crystal structures. Our method provides a way to characterise pores in transmembrane proteins and may even provide a starting point for discovering novel routes of therapeutic intervention in a number of important diseases. This software is freely available as source code from: http://bioinf.cs.ucl.ac.uk/downloads/memsat-svm/.
Figures
Figure 1
Prediction of pore-lining residues from sequence mapped onto native crystal structures. Homotetrameric calcium-gated potassium channel MthK (left, 1LNQ) bottom-up view; no false positives were predicted. Homodimeric molybdate transporter ModB2C2 (centre, 2ONK) top-down view. Homotetrameric AMPA-subtype glutamate receptor (3KG2, right) bottom-up view. The pore identified by Pore-Walker is shown by yellow spheres, while residues correctly predicted as pore-lining are shown in green (true positives), incorrectly predicted as pore-lining are shown in orange (false positives), correctly predicted as non-pore-lining are shown in grey (true negatives) and incorrectly predicted as non-pore-lining are shown in magenta (false negatives).
Figure 2
Prediction of pore-lining helices from sequence, mapped onto native crystal structures. Homotetrameric potassium channel Kir3.1 (left, 2QKS); both helices in each subunit are correctly classified. Monomeric 12 transmembrane helix multiple-drug resistance transporter NorM (right, 3MKT); 10 out of 12 helices are correctly classified–one false positive and one false negative are predicted. The pore identified by Pore-Walker is shown in yellow spheres, while helices correctly predicted as pore-lining are shown in green (true positives), incorrectly predicted as pore-lining are shown in orange (false positives), correctly predicted as non-pore-lining are shown in grey (true negatives) and incorrectly predicted as non-pore-lining are shown in magenta (false negatives). In both cases, all transmembrane helices were correctly predicted by MEMSAT-SVM.
Figure 3
Correctly predicted stoichiometries of three multimeric pore complexes. Bottom-up view of the heterodimeric maltose transporter malFG (left, 2R6G chain F), homotetrameric inward rectifier potassium channel KirBac1.1 (centre, 1P7B) and heterohexameric formate dehydrogenase (right, 1KQG chain B). Stoichiometry relates to the pore-lining membrane region only–extramembranous chains or those that do not line the pore are excluded. Complexes are coloured by chain, with the Pore-Walker predicted location of the pore shown by yellow spheres.
Similar articles
- PRIMSIPLR: prediction of inner-membrane situated pore-lining residues for alpha-helical transmembrane proteins.
Nguyen D, Helms V, Lee PH. Nguyen D, et al. Proteins. 2014 Jul;82(7):1503-11. doi: 10.1002/prot.24520. Epub 2014 Feb 18. Proteins. 2014. PMID: 24464816 - Transmembrane protein topology prediction using support vector machines.
Nugent T, Jones DT. Nugent T, et al. BMC Bioinformatics. 2009 May 26;10:159. doi: 10.1186/1471-2105-10-159. BMC Bioinformatics. 2009. PMID: 19470175 Free PMC article. - Predicting transmembrane helix packing arrangements using residue contacts and a force-directed algorithm.
Nugent T, Jones DT. Nugent T, et al. PLoS Comput Biol. 2010 Mar 19;6(3):e1000714. doi: 10.1371/journal.pcbi.1000714. PLoS Comput Biol. 2010. PMID: 20333233 Free PMC article. - Bridging the gap between structural bioinformatics and receptor research: the membrane-embedded, ligand-gated, P2X glycoprotein receptor.
Mager PP, Weber A, Illes P. Mager PP, et al. Curr Top Med Chem. 2004;4(16):1657-705. doi: 10.2174/1568026043387197. Curr Top Med Chem. 2004. PMID: 15579102 Review. - Emerging issues of connexin channels: biophysics fills the gap.
Harris AL. Harris AL. Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
Cited by
- Prediction and validation of the structural features of Ov58GPCR, an immunogenic determinant of Onchocerca volvulus.
Shey RA, Ghogomu SM, Njume FN, Gainkam LOT, Poelvoorde P, Mutesa L, Robert A, Humblet P, Munyampundu JP, Kamgno J, Lelubre C, Vanhamme L, Souopgui J. Shey RA, et al. PLoS One. 2018 Sep 26;13(9):e0202915. doi: 10.1371/journal.pone.0202915. eCollection 2018. PLoS One. 2018. PMID: 30256790 Free PMC article. - Diversity and conservation of plant small secreted proteins associated with arbuscular mycorrhizal symbiosis.
Hu XL, Zhang J, Kaundal R, Kataria R, Labbé JL, Mitchell JC, Tschaplinski TJ, Tuskan GA, Cheng ZM, Yang X. Hu XL, et al. Hortic Res. 2022 Feb 19;9:uhac043. doi: 10.1093/hr/uhac043. Online ahead of print. Hortic Res. 2022. PMID: 35184190 Free PMC article. - Large-scale ORF screening based on LC-MS to discover novel lncRNA-encoded peptides responding to ionizing radiation and microgravity.
Li W, Yu Y, Zhou G, Hu G, Li B, Ma H, Yan W, Pei H. Li W, et al. Comput Struct Biotechnol J. 2023 Oct 20;21:5201-5211. doi: 10.1016/j.csbj.2023.10.040. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37928948 Free PMC article. - Halocin H4 is activated through cleavage by halolysin HlyR4.
Chen S, Dai Y, Ke J, Luo Y, Wang C, Hao Y, Zhang A, Han J, Xiang H. Chen S, et al. Appl Environ Microbiol. 2024 Apr 17;90(4):e0228423. doi: 10.1128/aem.02284-23. Epub 2024 Mar 6. Appl Environ Microbiol. 2024. PMID: 38445904 Free PMC article. - Scalable web services for the PSIPRED Protein Analysis Workbench.
Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Buchan DW, et al. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W349-57. doi: 10.1093/nar/gkt381. Epub 2013 Jun 8. Nucleic Acids Res. 2013. PMID: 23748958 Free PMC article.
References
- Engel A, Walz T, Agre P. The aquaporin family of membrane water channels. Curr Opin Struct Biol. 1994;4:545–553. doi: 10.1016/S0959-440X(94)90217-8. - DOI
- Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 2000;68:425–458. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases