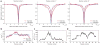Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy - PubMed (original) (raw)
Probing exchange kinetics and atomic resolution dynamics in high-molecular-weight complexes using dark-state exchange saturation transfer NMR spectroscopy
Nicolas L Fawzi et al. Nat Protoc. 2012.
Abstract
We present the protocol for the measurement and analysis of dark-state exchange saturation transfer (DEST), a novel solution NMR method for characterizing, at atomic resolution, the interaction between an NMR-'visible' free species and an NMR-'invisible' species transiently bound to a very high-molecular-weight (>1 MDa) macromolecular entity. The reduced rate of reorientational motion in the bound state that precludes characterization by traditional NMR methods permits the observation of DEST. (15)N-DEST profiles are measured on a sample comprising the dark state in exchange with an NMR-visible species; in addition, the difference (ΔR(2)) in (15)N transverse relaxation rates between this sample and a control sample comprising only the NMR-visible species is also obtained. The (15)N-DEST and ΔR(2) data for all residues are then fitted simultaneously to the McConnell equations for various exchange models describing the residue-specific dynamics in the bound state(s) and the interconversion rate constants. Although the length of the experiments depends strongly on sample conditions, approximately 1 week of NMR spectrometer time was sufficient for full characterization of samples of amyloid-β (Aβ) at concentrations of ~100 μM.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Figures
Figure 1
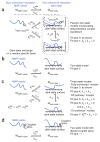
Summary of the kinetic models available in the DESTfit program. (a) Pseudo–two-state models in which the equilibrium between the NMR-visible and dark states is described by a single konapp and _k_off, connecting the free (MNMR-visible) and dark (Mdark-state) states of the exchanging species, whereas the dark state is partitioned into residue-specific equilibria of ensembles of tethered (Itethered) and direct-contact (Icontact) states for residue i, such that the concentration of INMR-visible = MNMR-visible for all i (i.e., M refers to the entire molecule, whereas I provides a description on a residue basis). (b) A two-state model, described by a single konapp and _k_off. (c) Three-state models in which two thermodynamically distinct dark states are populated, including fully connected (Fit types 3 and 4), ‘off-pathway’ (Fit types 5 and 6) and ‘on-pathway’ (Fit type 7) models. (d) A two-state model incorporating the effect of dipolar coupling between spins of different chemical shifts, most useful for analysis of 1H DEST data.
Figure 2
Screenshots of figures output by the DESTfit software. (a) Comparison of experimental (red) and calculated (blue) 15N-DEST profiles observed for Aβ40 at 260 μM for residues E3 (left), L17 (center) and N27 (right). (b) Comparison of experimental (red) and simulated (blue) Δ_R_2 for Aβ40 at 260 μM relative to Aβ40 at 50 μM. (c,d) Best-fit residue-specific R2tethered (c) and _K_3 (d), in which the error bars represent confidence intervals for one s.d. Calculated experimental DEST and Δ_R_2 profiles for the best-fit pseudo–two-state model with dark states comprising tethered and direct-contact ensembles, fit type 1 (blue solid lines) show significantly better agreement with experimental values compared with those for the best-fit simple two-state model, fit type 2 (blue dotted lines). Expt data, experimental data; freq, frequency.
Similar articles
- Decorrelating Kinetic and Relaxation Parameters in Exchange Saturation Transfer NMR: A Case Study of N-Terminal Huntingtin Peptides Binding to Unilamellar Lipid Vesicles.
Ceccon A, Clore GM, Tugarinov V. Ceccon A, et al. J Phys Chem B. 2018 Dec 13;122(49):11271-11278. doi: 10.1021/acs.jpcb.8b07112. Epub 2018 Sep 12. J Phys Chem B. 2018. PMID: 30156416 Free PMC article. - Characterizing methyl-bearing side chain contacts and dynamics mediating amyloid β protofibril interactions using ¹³C(methyl)-DEST and lifetime line broadening.
Fawzi NL, Libich DS, Ying J, Tugarinov V, Clore GM. Fawzi NL, et al. Angew Chem Int Ed Engl. 2014 Sep 22;53(39):10345-9. doi: 10.1002/anie.201405180. Epub 2014 Aug 11. Angew Chem Int Ed Engl. 2014. PMID: 25130489 Free PMC article. - Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR.
Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Fawzi NL, et al. Nature. 2011 Oct 30;480(7376):268-72. doi: 10.1038/nature10577. Nature. 2011. PMID: 22037310 Free PMC article. - NMR methods for exploring 'dark' states in ligand binding and protein-protein interactions.
Tugarinov V, Ceccon A, Clore GM. Tugarinov V, et al. Prog Nucl Magn Reson Spectrosc. 2022 Feb;128:1-24. doi: 10.1016/j.pnmrs.2021.10.001. Epub 2021 Nov 2. Prog Nucl Magn Reson Spectrosc. 2022. PMID: 35282867 Free PMC article. Review. - Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding.
Korzhnev DM, Kay LE. Korzhnev DM, et al. Acc Chem Res. 2008 Mar;41(3):442-51. doi: 10.1021/ar700189y. Epub 2008 Feb 15. Acc Chem Res. 2008. PMID: 18275162 Review.
Cited by
- Polyhydroxylated [60]fullerene binds specifically to functional recognition sites on a monomeric and a dimeric ubiquitin.
Zanzoni S, Ceccon A, Assfalg M, Singh RK, Fushman D, D'Onofrio M. Zanzoni S, et al. Nanoscale. 2015 Apr 28;7(16):7197-205. doi: 10.1039/c5nr00539f. Nanoscale. 2015. PMID: 25811293 Free PMC article. - NMR Lineshape Analysis of Intrinsically Disordered Protein Interactions.
Waudby CA, Christodoulou J. Waudby CA, et al. Methods Mol Biol. 2020;2141:477-504. doi: 10.1007/978-1-0716-0524-0_24. Methods Mol Biol. 2020. PMID: 32696373 Free PMC article. - Recent excitements in protein NMR: Large proteins and biologically relevant dynamics.
Chiliveri SC, Deshmukh MV. Chiliveri SC, et al. J Biosci. 2016 Dec;41(4):787-803. doi: 10.1007/s12038-016-9640-y. J Biosci. 2016. PMID: 27966496 Review. - Reversible Kinetic Trapping of FUS Biomolecular Condensates.
Chatterjee S, Kan Y, Brzezinski M, Koynov K, Regy RM, Murthy AC, Burke KA, Michels JJ, Mittal J, Fawzi NL, Parekh SH. Chatterjee S, et al. Adv Sci (Weinh). 2022 Feb;9(4):e2104247. doi: 10.1002/advs.202104247. Epub 2021 Dec 4. Adv Sci (Weinh). 2022. PMID: 34862761 Free PMC article. - Studies of Dynamic Binding of Amino Acids to TiO2 Nanoparticle Surfaces by Solution NMR and Molecular Dynamics Simulations.
Xue M, Sampath J, Gebhart RN, Haugen HJ, Lyngstadaas SP, Pfaendtner J, Drobny G. Xue M, et al. Langmuir. 2020 Sep 8;36(35):10341-10350. doi: 10.1021/acs.langmuir.0c01256. Epub 2020 Aug 26. Langmuir. 2020. PMID: 32693593 Free PMC article.
References
- Kay LE. Solution NMR spectroscopy of supra-molecular systems, why bother? A methyl-TROSY view. J Magn Reson. 2011;210:159–170. - PubMed
- Ishima R, Torchia DA. Accuracy of optimized chemical-exchange parameters derived by fitting CPMG R2 dispersion profiles when R20a≠R20b. J Biomol NMR. 2006;34:209–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources