The origin and evolution of mutations in acute myeloid leukemia - PubMed (original) (raw)
. 2012 Jul 20;150(2):264-78.
doi: 10.1016/j.cell.2012.06.023.
Timothy J Ley, Daniel C Link, Christopher A Miller, David E Larson, Daniel C Koboldt, Lukas D Wartman, Tamara L Lamprecht, Fulu Liu, Jun Xia, Cyriac Kandoth, Robert S Fulton, Michael D McLellan, David J Dooling, John W Wallis, Ken Chen, Christopher C Harris, Heather K Schmidt, Joelle M Kalicki-Veizer, Charles Lu, Qunyuan Zhang, Ling Lin, Michelle D O'Laughlin, Joshua F McMichael, Kim D Delehaunty, Lucinda A Fulton, Vincent J Magrini, Sean D McGrath, Ryan T Demeter, Tammi L Vickery, Jasreet Hundal, Lisa L Cook, Gary W Swift, Jerry P Reed, Patricia A Alldredge, Todd N Wylie, Jason R Walker, Mark A Watson, Sharon E Heath, William D Shannon, Nobish Varghese, Rakesh Nagarajan, Jacqueline E Payton, Jack D Baty, Shashikant Kulkarni, Jeffery M Klco, Michael H Tomasson, Peter Westervelt, Matthew J Walter, Timothy A Graubert, John F DiPersio, Li Ding, Elaine R Mardis, Richard K Wilson
Affiliations
- PMID: 22817890
- PMCID: PMC3407563
- DOI: 10.1016/j.cell.2012.06.023
The origin and evolution of mutations in acute myeloid leukemia
John S Welch et al. Cell. 2012.
Abstract
Most mutations in cancer genomes are thought to be acquired after the initiating event, which may cause genomic instability and drive clonal evolution. However, for acute myeloid leukemia (AML), normal karyotypes are common, and genomic instability is unusual. To better understand clonal evolution in AML, we sequenced the genomes of M3-AML samples with a known initiating event (PML-RARA) versus the genomes of normal karyotype M1-AML samples and the exomes of hematopoietic stem/progenitor cells (HSPCs) from healthy people. Collectively, the data suggest that most of the mutations found in AML genomes are actually random events that occurred in HSPCs before they acquired the initiating mutation; the mutational history of that cell is "captured" as the clone expands. In many cases, only one or two additional, cooperating mutations are needed to generate the malignant founding clone. Cells from the founding clone can acquire additional cooperating mutations, yielding subclones that can contribute to disease progression and/or relapse.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
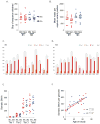
Figure 1. Coverage and number of validated single nucleotide variants (SNVs) by tier per genome
A. Gigabase pairs of sequence obtained for each genome assessed by whole genome sequencing. B. Mean number of reads obtained for each variant assessed per genome during validation. C and D. Number of tier 1, 2, 3, and total variants validated in each sample. E. Total number of validated SNVs by tier in M1 AML (red) versus M3 AML (blue) cases. F. Total number of validated SNVs per genome in non-redundant regions (tier 1, 2, and 3) versus age of patient; M1 AML (red), M3 AML (blue). The R2 for the combined 24 cases is 0.5, p < 0.0001.
Figure 2. Genomic distribution of somatic variants
A. Percentage of validated SNVs in each tier versus the number of genomic megabases occupied by that tier; M1 (red), M3 (blue). Average R2 M1: 0.999 (range: 0.998–1.000); M3: 0.999 (range: 0.998–1.000). B. Distribution of validated SNVs throughout the genome. Each point represents the genomic position of an SNV: X axis represents chromosomes in numerical order; Y axis represents base pair position on each chromosome in megabase pairs; M1 AML (red), M3 AML (blue). The large discontinuous regions represent regions of the reference genome without defined sequence. C. Average (circle), standard deviation (bars), and maximal (triangles) number of SNVs per megabase by chromosome in all 24 cases. D. Frequency of SNVs per megabase of non-tier 4 genomic space compared with the expected Poisson distribution if SNVs were randomly distributed across non-tier 4 space. E. Mutational spectrum of validated SNVs. Percentage of validated SNVs that occur in non-redundant regions of the genome (tiers 1, 2, and 3) with indicated nucleotide changes in M1 AML (red) and M3 AML (blue). F. Mutational spectrum as defined by tier of each event (tier 1, genic; tier 2, highly conserved and/or with regulatory potential; tier 3, unique in the genome, and not in tier 1 or tier 2, see Mardis et. al. 2009 for details).
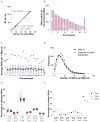
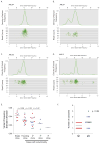
Figure 3. Clonality assessment in NK M1 AML and t(15;17)-positive M3 AML
A–D. Clusters of variants that occur with similar frequencies identify AML founding clones and subclones. Upper panels: probability density function using kernel density estimation of data in lower panels. The graphed line shows the relative probability of each variant allele frequency in the total sample. Lower panels: for each SNV, the frequency of the variant allele within the bone marrow sample is plotted versus the total number of sequencing reads covering the corresponding nucleotide position. Triangles indicate SNVs on the X chromosome. E. The standard deviation of the variant frequency for each cluster in all 24 cases: M1 AML (red); M3 AML (blue). F. Number of clusters identified by kernel density estimation in M1 AML (red) and M3 AML (blue).
Figure 4. Background mutations in normal HSPCs accumulate as a function of age
A. Number of validated SNVs per exome identified in each of 3 clones that were derived from individual hematopoietic stem/progenitor cells from 7 anonymous, healthy donors with normal blood counts. B. Clustering of mutations by clone. The Y-axis represents the analyzed clones from each donor. The X-axis represents the genes with mutations in each clone (red). Note that the two clones with ZFHX4 variants contained unique mutations (Q1660K vs. P3497A). C. Mutational spectrum in normal donor exomes (81 validated variants) vs. 24 tier 1 AML genomes (343 validated variants).
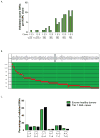
Figure 5. Recurrently mutated genes in M1 and M3 AML
A. 108 total cases of M1 and M3 AML were screened for somatic variants in the 284 genes identified by WGS and in 53 genes previously reported to be mutated in AML. Mutations with translational consequences that occur in at least three unique genomes are represented in red. FAB subtype indicated above graph. Overall survival is indicated by red (≤ 12 months), pink (12–24 months), and white (> 24 months) bars above graph. B. The total number of validated genic variants per genome identified by WGS in 12 NK M1 AML cases (red) and 12 M3 AML cases (blue), including PML-RARA. C. The total number of recurrently mutated genes per genome identified in 12 NK M1 AML cases (red) and 12 M3 AML cases (blue), including PML-RARA. Only mutations with translational consequences were assessed. D. Total number of recurrently mutated genes per genome identified in 65 M1 AML cases (red) and 43 M3 AML cases (blue), including PML-RARA. Only mutations with translational consequences were assessed. E. Somatic variants in cohesin complex genes (SMC3, SMC1A, STAG2, and RAD21) identified in 199 cases of AML (red). Additional mutations with translational consequences in highly recurrent genes are also shown for each genome (red).
Figure 6. Pathway analysis of key mutations in the 24 sequenced genomes
Mutations identified in Table 1 were individually curated in Pubmed for biological function. Functional categories represented by 2 or fewer mutated genes were excluded. Mutations in NPM1, DNA methylation, and cohesin genes occurred only in the M1 cases, suggesting that they may substitute for PML-RARA as initiating events, or do not productively cooperate with PML-RARA. Mutations in ubiquitin pathway genes occurred more often in M3 AML. Genes representing other functional categories were mutated in both M1 and M3 AML cases.
Figure 7. Integrated model for the origin of driver and passenger mutations during AML evolution
Hematopoietic stem/progenitor cells (HSPCs, shown in green) are long-lived cells that accumulate random, benign background mutations as a function of age. Based on the fact that HSPCs from normal, healthy donors accumulate cell-specific, age-dependent mutations, and that the total mutational burden in AML genomes is related to age, we suggest that the largest numbers of mutations in AML genomes are variants that were preexisting in the cell that received the initiating event. “X” represents these background mutations in the HSPCs, and we estimate that X may range between 100 and 1000 events, depending on age. The initiating mutations, which are drivers, are different for M1 AML cases and M3 AML cases. The initiating event provides an advantage for the affected cell and clonal expansion ensues, so that all of the preexisting mutations are “captured” by cloning. Each progression event gives the expanding clone an additional advantage; based on the data in this paper, we suggest that 1–5 events contribute to progression in most cases of AML. Each progression mutation is expected to capture all mutations that occurred between the initiating event and the progression event (designated as “Y” in the yellow cell). This number would be affected by the time between these events, and the mutation rate in these cells. Although this number is unknown at present, the analysis of clonal progression of secondary AML (Walter et al., 2012) suggests that each progression event may capture dozens to hundreds of mutations. Clonal outgrowth of cells with appropriate progression events results in AML, which is dominated by the “founding” AML clone, designated in red. When this clone is sequenced, the driver mutations (1–6 events) and all associated passenger mutations (hundreds of events) would all be present in all cells within the sample, which is what we observe. Subclones arise from the founding AML clone by acquiring a small number of additional mutations that confer an advantage to that cell, along with any additional background mutations that may have occurred in the interim (represented as “Z”). In this study, the average number of mutations captured in subclones was 40 (range 6–110).
Similar articles
- Clonal evolution of acute leukemia genomes.
Jan M, Majeti R. Jan M, et al. Oncogene. 2013 Jan 10;32(2):135-40. doi: 10.1038/onc.2012.48. Epub 2012 Feb 20. Oncogene. 2013. PMID: 22349821 Free PMC article. Review. - Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, Shannon WD, Payton JE, Kulkarni S, Westervelt P, Walter MJ, Graubert TA, Mardis ER, Wilson RK, DiPersio JF. Ding L, et al. Nature. 2012 Jan 11;481(7382):506-10. doi: 10.1038/nature10738. Nature. 2012. PMID: 22237025 Free PMC article. - Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia.
Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. Jan M, et al. Sci Transl Med. 2012 Aug 29;4(149):149ra118. doi: 10.1126/scitranslmed.3004315. Sci Transl Med. 2012. PMID: 22932223 Free PMC article. - Clonal evolution of acute myeloid leukemia highlighted by latest genome sequencing studies.
Zhang X, Lv D, Zhang Y, Liu Q, Li Z. Zhang X, et al. Oncotarget. 2016 Sep 6;7(36):58586-58594. doi: 10.18632/oncotarget.10850. Oncotarget. 2016. PMID: 27474172 Free PMC article. Review. - Monitoring of clonal evolution of double C-KIT exon 17 mutations by Droplet Digital PCR in patients with core-binding factor acute myeloid leukemia.
Tan Y, Liu Z, Wang W, Zhu G, Guo J, Chen X, Zheng C, Xu Z, Chang J, Ren F, Wang H. Tan Y, et al. Leuk Res. 2018 Jun;69:89-93. doi: 10.1016/j.leukres.2018.04.013. Epub 2018 Apr 22. Leuk Res. 2018. PMID: 29705537
Cited by
- DNA methylation drives hematopoietic stem cell aging phenotypes after proliferative stress.
Yanai H, McNeely T, Ayyar S, Leone M, Zong L, Park B, Beerman I. Yanai H, et al. Geroscience. 2024 Oct 11. doi: 10.1007/s11357-024-01360-4. Online ahead of print. Geroscience. 2024. PMID: 39390312 - Recent progress in AML with recurrent genetic abnormalities.
Ishikawa Y. Ishikawa Y. Int J Hematol. 2024 Nov;120(5):525-527. doi: 10.1007/s12185-024-03848-3. Epub 2024 Oct 1. Int J Hematol. 2024. PMID: 39352624 Review. - Developmental interplay between transcriptional alterations and a targetable cytokine signaling dependency in pediatric ETO2::GLIS2 leukemia.
Alonso-Pérez V, Galant K, Boudia F, Robert E, Aid Z, Renou L, Barroca V, Devanand S, Babin L, Rouiller-Fabre V, Moison D, Busso D, Piton G, Metereau C, Abermil N, Ballerini P, Hirsch P, Haddad R, Martinovic J, Petit A, Lapillonne H, Brunet E, Mercher T, Pflumio F. Alonso-Pérez V, et al. Mol Cancer. 2024 Sep 20;23(1):204. doi: 10.1186/s12943-024-02110-y. Mol Cancer. 2024. PMID: 39304903 Free PMC article. - Glutamine metabolism-related genes predict the prognostic risk of acute myeloid leukemia and stratify patients by subtype analysis.
Zhou J, Zhang N, Zuo Y, Xu F, Cheng L, Fu Y, Yang F, Shu M, Zhou M, Zou W, Zhang S. Zhou J, et al. Hereditas. 2024 Sep 19;161(1):35. doi: 10.1186/s41065-024-00338-8. Hereditas. 2024. PMID: 39300580 Free PMC article. - The Clonal Hematopoiesis-associated Gene Srcap Plays an Essential Role in Hematopoiesis.
Wong TN, Mychalowych A, Feldpausch ER, Carson A, Karpova D, Link DC. Wong TN, et al. bioRxiv [Preprint]. 2024 Aug 19:2024.08.16.607812. doi: 10.1101/2024.08.16.607812. bioRxiv. 2024. PMID: 39229096 Free PMC article. Preprint.
References
- Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nat Med. 1996;2:190–197. - PubMed
- Abkowitz JL, Golinelli D, Harrison DE, Guttorp P. In vivo kinetics of murine hemopoietic stem cells. Blood. 2000;96:3399–3405. - PubMed
- Allford S, Grimwade D, Langabeer S, Duprez E, Saurin A, Chatters S, Walker H, Roberts P, Rogers J, Bain B, et al. Identification of the t(15;17) in AML FAB types other than M3: evaluation of the role of molecular screening for the PML/RARalpha rearrangement in newly diagnosed AML. The Medical Research Council (MRC) Adult Leukaemia Working Party. Br J Haematol. 1999;105:198–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA91842/CA/NCI NIH HHS/United States
- P01 CA101937/CA/NCI NIH HHS/United States
- R01 CA083962/CA/NCI NIH HHS/United States
- K99 HL103975/HL/NHLBI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- R00 HL103975/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases