Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons - PubMed (original) (raw)
Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons
H Patil et al. Cell Death Dis. 2012.
Abstract
The retinitis pigmentosa GTPase regulator (RPGR) and nephrocystin-4 (NPHP4) comprise two key partners of the assembly complex of the RPGR-interacting protein 1 (RPGRIP1). Mutations in RPGR and NPHP4 are linked to severe multisystemic diseases with strong retinal involvement of photoreceptor neurons, whereas those in RPGRIP1 cause the fulminant photoreceptor dystrophy, Leber congenital amaurosis (LCA). Further, mutations in Rpgrip1 and Nphp4 suppress the elaboration of the outer segment compartment of photoreceptor neurons by elusive mechanisms, the understanding of which has critical implications in uncovering the pathogenesis of syndromic retinal dystrophies. Here we show RPGRIP1 localizes to the photoreceptor connecting cilium (CC) distally to the centriole/basal body marker, centrin-2 and the ciliary marker, acetylated-α-tubulin. NPHP4 abuts proximally RPGRIP1, RPGR and the serologically defined colon cancer antigen-8 (SDCCAG8), a protein thought to partake in the RPGRIP1 interactome and implicated also in retinal-renal ciliopathies. Ultrastructurally, RPGRIP1 localizes exclusively throughout the photoreceptor CC and Rpgrip1(nmf247) photoreceptors present shorter cilia with a ruffled membrane. Strikingly, Rpgrip1(nmf247) mice without RPGRIP1 expression lack NPHP4 and RPGR in photoreceptor cilia, whereas the SDCCAG8 and acetylated-α-tubulin ciliary localizations are strongly decreased, even though the NPHP4 and SDCCAG8 expression levels are unaffected and those of acetylated-α-tubulin and γ-tubulin are upregulated. Further, RPGRIP1 loss in photoreceptors shifts the subcellular partitioning of SDCCAG8 and NPHP4 to the membrane fraction associated to the endoplasmic reticulum. Conversely, the ciliary localization of these proteins is unaffected in glomeruli or tubular kidney cells of Rpgrip1(nmf247), but NPHP4 is downregulated developmentally and selectively in kidney cortex. Hence, RPGRIP1 presents cell type-dependent pathological effects crucial to the ciliary targeting and subcellular partitioning of NPHP4, RPGR and SDCCAG8, and acetylation of ciliary α-tubulin or its ciliary targeting, selectively in photoreceptors, but not kidney cells, and these pathological effects underlie photoreceptor degeneration and LCA.
Figures
Figure 1
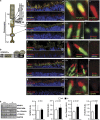
RPGRIP1_α_ determines distinct ciliary localizations of proteins in mouse photoreceptor neurons. (a) Schematic diagram of a rod photoreceptor with its ciliary region connecting the inner (IS) and outer segment (OS) compartments depicted within the circle (left). Structural organization of the schematic and amplified ciliary region is noted on the right. (b) Overall primary structure of RPGRIP1_α_ with its domains, protein kinase C conserved region 2 (C2) and RPGR-interacting domain (RID), and its interacting partners, NPHP4 and RPGR, and examined by this study. (c–g) Left and right panels, respectively, are low- and high-magnification images of distal regions of P12 retinas showing: localization of RPGRIP1_α_ in the connecting cilium and juxtaposed distally to the centriolar/basal body marker, centrin-2, whose localization in the basal body is not affected by loss of RPGRIP1_α_ in Rpgrip1 nmf247 (c); ciliary localization of RPGRIP1_α_ distal of and abutting acetylated α_-tubulin and whose ciliary signal is strongly decreased in Rpgrip1 nmf247 (d); ciliary localization of RPGRIP1_α distal to NPHP4, whose ciliary localization is abolished in Rpgrip1 nmf247 (e); localization of RPGR at the connecting cilium distal to NPHP4, the ciliary localization of both proteins are abolished in Rpgrip1 nmf247 (f); NPHP4 ciliary localization abutting proximally SDCCAG8, whose ciliary localization is strongly reduced in Rpgrip1 nmf247 (g). Note the localization of NPHP4 extends into the inner segments to the striated rootlets. (h) Quantitation of ciliary proteins from immunoblots of retinal homogenates (left). NPHP4 and SDCCAG8 levels do not change between wild type and Rpgrip1 nmf247, whereas acetylated _α_-tubulin and _γ_-tubulin are upregulated in Rpgrip1 nmf247. Bars represent the mean±S.D. (_n_=4). P<0.05 is considered significant. Low- and high-magnification scale bars: 10 and 0.5 _μ_m; +/+, wild type; −/− Rpgrip1 nmf247; AU, arbitrary units; AX, axoneme; BB, basal body; CC/TZ, connecting cilium/transition zone; DC, daughter centriole; Nuc, nucleus; SR, striated rootlet; Syn, synapse. Dash arrows point to locations of schematic regions in photoreceptor neurons in the retinal sections
Figure 2
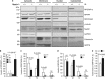
Loss of RPGRIP1_α_ expression promotes the accumulation of SDCCAG8 and NPHP4 in the retinal membrane subcellular fraction and ER stress. (a) Immunoblots of cytosolic (C), membrane (M), nuclear (N) and cytoskeletal (Ck) subcellular fractions of P14 retinas of wild-type (+/+) and Rpgrip1 nmf247 (−/−) mice with antibodies against the ciliary proteins, RPGRIP1, NPHP4, SDCCAG8, and the subcellular makers, γ_-tubulin (ciliary/cytoskeleton), Nup62 (pan-subcellular marker), GAPDH (cytosol), calreticulin (ER) and GRP78 (ER and cytosolic stress marker). RPGRIP1_α is mostly in the cytoskeletal fraction, whereas NPHP4 and SDCCAG8 are found mostly in the cytosolic and membrane fractions. The low levels of localization of these components in the nuclear fractions likely represents carry-over of ER membranes connected to the nuclear membrane. In Rpgrip1 nmf247 mice, there is upregulation of careticulin and GRP78. (b) Quantitation of the relative levels of RPGRIP1_α_ between subcellular retinal fractions. RPGRIP1_α_ is predominant in the cytoskeletal (Ck) fraction of +/+ mice (n_=4, −/− n_=5, +/+). (c) Quantitation of the relative levels of SDCCAG8 (c) and NPHP4 (d) between subcellular retinal fractions of wild-type (+/+) and Rpgrip1 nmf247 (−/−) mice. SDCCAG8 is localized predominantly in the cytosolic (C) and membrane (M) fractions of +/+ mice, whereas its localization increases and decreases in the membrane (M), and cytosolic (C) and nuclear (N) fractions, respectively, of Rpgrip1 nmf247 mice (c). NPHP4 is localized predominantly in the cytosolic (C) and membrane (M) fractions of +/+ mice, whereas its localization increases and decreases in the membrane (M) and nuclear (N) fractions, respectively, of Rpgrip1 nmf247 mice (d). (e) Total levels of GRP78 between retinas of wild-type (+/+) and Rpgrip1 nmf247 (−/−) mice. There is a ∼1.5-fold increase of GRP78 in retinas of Rpgrip1 nmf247 (−/−) mice. Bars represent the mean±S.D. P<0.05 was considered significant (Student's _t_-test). n_=4 (−/−) and n_=5 (+/+) in (b), (c) and (d); _n_=4 (−/−, +/+) in (e). Nup62, nucleoporin 62; GAPDH, glyceraldehyde 3-phosphate dehydrogenase
Figure 3
The ciliary/centriolar localization of _α_-tubulin, NPHP4 and SDCCAG8 is unaffected in P21 kidney cells of Rpgrip1 nmf247 mice. Images are low and high magnifications showing: (a) partial colocalization of acetylated _α_-tubulin and SDCCAG8 at the centrioles of glomeruli (Gl) and surrounding kidney tubular cells of wild-type and Rpgrip1 nmf247 mice, the centriolar colocalization is not affected in Rpgrip1 nmf247; (b) NPHP4 and RPGR subcellular distributions varied between glomeruli and tubular cells, their ciliary (middle panel) or centriolar localizations do not overlap (lower panel) in wild-type or Rpgrip1 nmf247 mice; (c) NPHP4 does not colocalize with SDCCAG8 in most glomeruli (Gl) cells of wild type and Rpgrip1 nmf247 (upper panels), whereas in tubular cells, the SDCCAG8-stained basal foot (BF) and cap (BC) appendages of the basal body flank laterally the NPHP4-stained axoneme and basal body (lower panel, amplified region of boxed area of upper panel). The partial colocalization of SDCCAG8 and NPHP4 is not affected in Rpgrip1 nmf247. (d) Schematic diagram of a cilium noted on the right. (e) Quantitation of ciliary proteins from immunoblots of medulla and cortex homogenates of kidneys of P12 and P21of age (left). NPHP4 and _γ_-tubulin levels decrease and increase, respectively, in the cortex but not medulla of P21 kidneys. P12 kidneys lack changes in ciliary protein levels between wild type and Rpgrip1 nmf247. Bars represent the mean±S.D. P<0.05 is considered significant; P12, _n_=4; P21, _n_=3. +/+, Wild type; −/−, Rpgrip1 nmf247; AU, arbitrary units; AX, axoneme; BB, basal body; DC, daughter centriole; TZ, transition zone; SR, striated rootlet
Figure 4
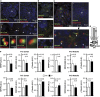
The apical targeting of rhodopsin and M-opsin is unaffected in Rpgrip1 nmf247 photoreceptors. (a) Localization of rhodopsin in rod photoreceptors of wild-type (+/+, left panels) and Rpgrip1 nmf247 (−/−, right panels) at P12 and P14 of age with two different antibodies against rhodopsin, 1D4 (upper panels) and RET-P1 antibodies (lower panels). Note, the lack of mislocalization or accumulation of rhodopsin in the cell bodies of photoreceptors with any of the antibodies and the correct polarized targeting of rhodopsin to the apical end of the inner segments of rod photoreceptors of Rpgrip1 nmf247 mice. In comparison to the 1D4 antibody, the RET-P1 antibody detects rhodopsin also in the photoreceptor nuclear layer (ONL) of wild-type and Rpgrip1 nmf247 mice. Images were acquired under the same exact acquisition parameters. Duplicated regions stained with DAPI of retinal sections shown are also displayed for reference to the localization of the cell bodies (ONL) of photoreceptors. (b) Qualitative (upper panel) and quantitative (lower panel) immunoblot analyses of rhodopsin levels in retinal homogenates of wild-type and Rpgrip1 nmf247 mice of P12.5 of age. Bars represent the mean±S.D. (n=4). P<0.05 is considered significant. (c) Localization of M-opsin in cone photoreceptors of wild-type (left panel) and Rpgrip1 nmf247 mice (right panels) at P14 of age. Note, the lack of mislocalization or accumulation of M-opsin in the cell bodies of cone photoreceptors and the correct polarized targeting of M-opsin to the apical end of the inner segments of cone photoreceptors of Rpgrip1 nmf247 mice. The number of M-cone photoreceptors is also decreased in Rpgrip1 nmf247 mice. Images were acquired under the same exact acquisition parameters. Duplicated regions stained with DAPI of retinal sections shown are also displayed for reference to the localization of the cell bodies (ONL) of photoreceptors. Scale bars: 20 _μ_m in all panels, except in panels of P14 of age with 1D4 staining, where scale bar is 10 _μ_m. +/+, Wild type; −/−, Rpgrip1 nmf247; AU, arbitrary units; DAPI, 4',6-diamidino-2-phenylindole; IS, inner segment of photoreceptors; ONL, outer nuclear layer; OS, outer segment of photoreceptors
Figure 5
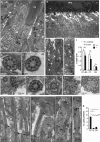
Ultrastructural ciliary changes and localization of RPGRIP1_α_ in photoreceptors. Transmission electron micrographs of photoreceptors and their cilia in P12.5 wild type (+/+) (a–c) and Rpgrip1 nmf247 (−/−) mice (d–k). (a) Longitudinal section of ciliary regions of rod photoreceptors. (b–d) Transverse sections of the connecting cilium at the distal level of transition zone with an amorphous disk structure and ciliary necklace. (d–f) Low and high magnifications of longitudinal sections of photoreceptors of Rpgrip1 nmf247 mice. Note, Rpgrip1 nmf247 cilia are naked of outer segments. (g–k) Proximal to distal transverse sections of the connecting cilium depicting the ruffling of the ciliary membrane. (l) Quantitative analyses of the length of basal bodies (BB) and connecting cilia (CC) between wild-type and Rpgrip1 nmf247 photoreceptors with the latter having shorter cilia. Bars represent the mean±S.D. (n_=21,−/−; n_=11_, +/+_ from three mice of each genotype). P<0.05 was considered significant (Student's t_-test). Only the longest cilia observed in Rpgrip1 nmf247 mice from the sampling of a large number of cilia were included for quantitation analysis. (m–p) Representative ultrastructural images of the restricted immunogold localization of RPGRIP1_α throughout the connecting cilium (CC) of photoreceptors. RPGRIP1_α_ was excluded from the basal body (arrow) and outer segment/axoneme of photoreceptors. RPGRIP1_α_ localization was prominent beneath the ciliary membrane (e.g. m, p) and at the proximal and distal regions of the connecting cilium (e.g. n–p). (q) Quantitation of immunogold particles of RPGRIP1_α_ in the connecting cilium (CC), inner (IS) and outer segments (OS) of photoreceptors. The majority of immunogold particles are restricted to the connecting cilium (CC). Bars represent the mean±S.D. (_n_=22) P<0.05 was considered significant (Student's _t_-test). White arrow, connecting cilium; black arrow, basal body; black arrowhead, proximal (daughter) centriole; white arrowhead, striated rootlets. OS, outer segments of rod photoreceptors; PC, periciliary ridge; RPE, retina pigment epithelium
Similar articles
- Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina.
Eblimit A, Nguyen TM, Chen Y, Esteve-Rudd J, Zhong H, Letteboer S, Van Reeuwijk J, Simons DL, Ding Q, Wu KM, Li Y, Van Beersum S, Moayedi Y, Xu H, Pickard P, Wang K, Gan L, Wu SM, Williams DS, Mardon G, Roepman R, Chen R. Eblimit A, et al. Hum Mol Genet. 2015 Mar 15;24(6):1584-601. doi: 10.1093/hmg/ddu573. Epub 2014 Nov 14. Hum Mol Genet. 2015. PMID: 25398945 Free PMC article. - Replacement gene therapy with a human RPGRIP1 sequence slows photoreceptor degeneration in a murine model of Leber congenital amaurosis.
Pawlyk BS, Bulgakov OV, Liu X, Xu X, Adamian M, Sun X, Khani SC, Berson EL, Sandberg MA, Li T. Pawlyk BS, et al. Hum Gene Ther. 2010 Aug;21(8):993-1004. doi: 10.1089/hum.2009.218. Hum Gene Ther. 2010. PMID: 20384479 Free PMC article. - Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations.
Roepman R, Letteboer SJ, Arts HH, van Beersum SE, Lu X, Krieger E, Ferreira PA, Cremers FP. Roepman R, et al. Proc Natl Acad Sci U S A. 2005 Dec 20;102(51):18520-5. doi: 10.1073/pnas.0505774102. Epub 2005 Dec 9. Proc Natl Acad Sci U S A. 2005. PMID: 16339905 Free PMC article. - RPGRIP1 is mutated in Leber congenital amaurosis: a mini-review.
Koenekoop RK. Koenekoop RK. Ophthalmic Genet. 2005 Dec;26(4):175-9. doi: 10.1080/13816810500374441. Ophthalmic Genet. 2005. PMID: 16352478 Review. - RPGR-containing protein complexes in syndromic and non-syndromic retinal degeneration due to ciliary dysfunction.
Murga-Zamalloa CA, Swaroop A, Khanna H. Murga-Zamalloa CA, et al. J Genet. 2009 Dec;88(4):399-407. doi: 10.1007/s12041-009-0061-7. J Genet. 2009. PMID: 20090203 Free PMC article. Review.
Cited by
- The Role of the Microglial Cx3cr1 Pathway in the Postnatal Maturation of Retinal Photoreceptors.
Jobling AI, Waugh M, Vessey KA, Phipps JA, Trogrlic L, Greferath U, Mills SA, Tan ZL, Ward MM, Fletcher EL. Jobling AI, et al. J Neurosci. 2018 May 16;38(20):4708-4723. doi: 10.1523/JNEUROSCI.2368-17.2018. Epub 2018 Apr 18. J Neurosci. 2018. PMID: 29669747 Free PMC article. - The Genetic and Endoplasmic Reticulum-Mediated Molecular Mechanisms of Primary Open-Angle Glaucoma.
Rozpędek-Kamińska W, Wojtczak R, Szaflik JP, Szaflik J, Majsterek I. Rozpędek-Kamińska W, et al. Int J Mol Sci. 2020 Jun 11;21(11):4171. doi: 10.3390/ijms21114171. Int J Mol Sci. 2020. PMID: 32545285 Free PMC article. Review. - NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone.
Awata J, Takada S, Standley C, Lechtreck KF, Bellvé KD, Pazour GJ, Fogarty KE, Witman GB. Awata J, et al. J Cell Sci. 2014 Nov 1;127(Pt 21):4714-27. doi: 10.1242/jcs.155275. Epub 2014 Aug 22. J Cell Sci. 2014. PMID: 25150219 Free PMC article. - Spata7 is a retinal ciliopathy gene critical for correct RPGRIP1 localization and protein trafficking in the retina.
Eblimit A, Nguyen TM, Chen Y, Esteve-Rudd J, Zhong H, Letteboer S, Van Reeuwijk J, Simons DL, Ding Q, Wu KM, Li Y, Van Beersum S, Moayedi Y, Xu H, Pickard P, Wang K, Gan L, Wu SM, Williams DS, Mardon G, Roepman R, Chen R. Eblimit A, et al. Hum Mol Genet. 2015 Mar 15;24(6):1584-601. doi: 10.1093/hmg/ddu573. Epub 2014 Nov 14. Hum Mol Genet. 2015. PMID: 25398945 Free PMC article. - The Role of RPGR and Its Interacting Proteins in Ciliopathies.
Patnaik SR, Raghupathy RK, Zhang X, Mansfield D, Shu X. Patnaik SR, et al. J Ophthalmol. 2015;2015:414781. doi: 10.1155/2015/414781. Epub 2015 Jun 1. J Ophthalmol. 2015. PMID: 26124960 Free PMC article. Review.
References
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. - PubMed
- Besharse JC, Horst CJ.The photoreceptor connecting cilium, a model for the transition zoneIn: Bloodgood RA (ed)Ciliary and Flagellar Membranes New York, NY; 1990389–417.
- Hildebrandt F, Zhou W. Nephronophthisis-associated ciliopathies. J Am Soc Nephrol. 2007;18:1855–1871. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 EY005722/EY/NEI NIH HHS/United States
- EY019492/EY/NEI NIH HHS/United States
- GM083165/GM/NIGMS NIH HHS/United States
- 2P30-EY005722/EY/NEI NIH HHS/United States
- R01 GM083165/GM/NIGMS NIH HHS/United States
- R01 EY019492/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials