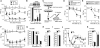IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype - PubMed (original) (raw)
IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype
Takahiro Masuda et al. Cell Rep. 2012.
Abstract
Microglia become activated by multiple types of damage in the nervous system and play essential roles in neuronal pathologies. However, how microglia transform into reactive phenotypes is poorly understood. Here, we identify the transcription factor interferon regulatory factor 8 (IRF8) as a critical regulator of reactive microglia. Within the spinal cord, IRF8 expression was normally low; however, the expression was markedly upregulated in microglia, but not in neurons or astrocytes, after peripheral nerve injury (PNI). IRF8 overexpression in cultured microglia promoted the transcription of genes associated with reactive states; conversely, IRF8 deficiency prevented these gene expressions in the spinal cord following PNI. Furthermore, IRF8-deficient mice were resistant to neuropathic pain, a common sequela of PNI, and transferring IRF8-overexpressing microglia spinally to normal mice produced pain. Therefore, IRF8 may activate a program of gene expression that transforms microglia into a reactive phenotype. Our findings provide a newly observed mechanism for microglial activation.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
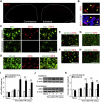
Figure 1. PNI Induces IRF8 Upregulation Exclusively in Microglia in the Spinal Cord
(A) Visualization of IRF8 protein in the dorsal spinal cord 3 days after PNI. (B) Nuclear localization of IRF8. (C–H) Double immunolabeling of IRF8 with Iba1 (C), OX-42 (D), GFAP (E), NeuN (F), MAP2 (G), and NF200 (H). (I) Real-time PCR analysis of Irf8 mRNA in WT mouse spinal cord before (Naive) and after PNI. Values represent the relative ratio of Irf8 mRNA (normalized to Gapdh mRNA) to the contralateral side of naive mice (n = 6; **p < 0.01). (J) Western blot analysis of IRF8 protein in the spinal cords of WT mice before (Naive) and after PNI. (K) A histogram of the relative band density ratio of IRF8 (normalized to β-actin) to the contralateral side of naive mice at each time point (n = 5; **p < 0.01). Values are means ± SEM for all groups. Scale bars: 100 μm (A), 10 μm (B), 50 μm (C–H). See also Figure S1.
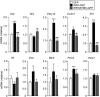
Figure 2. Forced Expression of IRF8 in Microglia Promotes Gene Transcription
Real-time PCR analysis of the mRNA of genes of interest in cultured microglia 72 hr after transduction. Values represent the relative ratio of mRNA (of tested genes, normalized to Gapdh mRNA) to control microglia with GFP alone (n = 5–7, *p < 0.05, ***p < 0.001 versus GFP; #p < 0.05, ##p < 0.01, ###p < 0.001 versus IRF8-GFP). Values are the mean ± SEM for all groups. See also Figure S2.
Figure 3. IRF8 Is Required for Microglial Gene Expression, but Not for Microglial Proliferation, in the Spinal Cord after PNI
(A) Real-time PCR analysis of mRNAs of microglial genes in the spinal cords of WT and _Irf8_–/– mice 7 days after PNI. Values represent the relative ratio of mRNA (of tested genes, normalized to Gapdh mRNA) to the contralateral side of WT mice. C, contralateral; I, ipsilateral. (n = 6–7, *p < 0.05, **p < 0.01, ***p < 0.001). (B) Double immunofluorescence for Ki-67 or p-HisH3 (green) and OX-42 (red) in the ipsilateral dorsal horn of WT or _Irf8_–/– mice 2 days after PNI. A histogram of the numbers of Ki-67+OX-42+ or p-HisH3+OX-42+ cells in the ipsilateral (I) and contralateral (C) dorsal horn (n = 3, ***p < 0.001). (C) OX-42 (red) and Iba1 (green) immunofluorescence in the spinal cord of WT and _Irf8_–/– mice 14 days after PNI. Values are the mean ± SEM for all groups. Scale bars: 50 μm (B), 200 μm (C), 50 μm (C, insets). See also Figure S3.
Figure 4. Microglial IRF8 Is Necessary for Abnormal Pain Hypersensitivity Caused by PNI
(A) PWT of _Irf8_–/– and WT mice before (Pre) and after PNI (n = 4; *p < 0.05, **p < 0.01 versus Pre; #p < 0.05, ##p < 0.01, ###p < 0.001 versus the ipsilateral side of WT mice). C, contralateral; I, ipsilateral. (B) Reversal of PNI-induced allodynia by intrathecal administration of IRF8 siRNA (20 pmol) once a day for 2 days (on 5 and 6 days post-PNI) in WT mice (n = 3–6, *p < 0.05). Upper, representative immunoblots of IRF8 and β-actin proteins in the spinal cords of mice treated with control and IRF8 siRNAs on day 7 post-PNI. (C) Experimental protocol. (D) PWT of WT mice intrathecally administered with cultured microglia overexpressing either GFP, IRF8–GFP or IRF8(K79E)–GFP (n = 5–6, **p < 0.01 versus Pre; #p < 0.05 versus WT mice with GFP microglia; §p < 0.05, §§p < 0.01 versus WT mice with IRF8–GFP microglia). (E) Allodynia by IRF8-transduced cultured microglia was prevented by preincubating microglia with a cocktail of IL-1β neutralizing antibody (5 μg) and CatS inhibitor (5 pmol) for 15 min before the intrathecal injection of microglia (n = 6–7, *p < 0.05, **p < 0.01 versus Pre; #p < 0.05, ###p < 0.001 versus IRF8–GFP/ control group). (F) PWT of WT and _Irf8_–/– mice before (Pre) and after intraplantar CFA injection (n = 5, **p < 0.01 versus Pre). (G) Hot-plate test. Values represent the latency (s) for animals to lick their hindpaws or jump (n = 6). (H) Tail-flick test. Values represent the latency (s) to flick their tail from the heat source (n = 4). (I–K) Capsaicin (I) and formalin (J and K) test. Values are the duration (s) of nociceptive behaviors (I: n = 6, J: n = 8) (**p < 0.01 versus WT mice). (K) Total duration (s) of nociceptive behaviors for 0–5 min (first phase) and for 10–60 min (second phase). Values are the mean ± SEM for all groups. See also Figure S4.
Similar articles
- Interferon regulatory factor 8/interferon consensus sequence binding protein is a critical transcription factor for the physiological phenotype of microglia.
Horiuchi M, Wakayama K, Itoh A, Kawai K, Pleasure D, Ozato K, Itoh T. Horiuchi M, et al. J Neuroinflammation. 2012 Sep 28;9:227. doi: 10.1186/1742-2094-9-227. J Neuroinflammation. 2012. PMID: 23020843 Free PMC article. - Transcription factor IRF5 drives P2X4R+-reactive microglia gating neuropathic pain.
Masuda T, Iwamoto S, Yoshinaga R, Tozaki-Saitoh H, Nishiyama A, Mak TW, Tamura T, Tsuda M, Inoue K. Masuda T, et al. Nat Commun. 2014 May 13;5:3771. doi: 10.1038/ncomms4771. Nat Commun. 2014. PMID: 24818655 Free PMC article. - Transcription factor IRF1 is responsible for IRF8-mediated IL-1β expression in reactive microglia.
Masuda T, Iwamoto S, Mikuriya S, Tozaki-Saitoh H, Tamura T, Tsuda M, Inoue K. Masuda T, et al. J Pharmacol Sci. 2015 Aug;128(4):216-20. doi: 10.1016/j.jphs.2015.08.002. Epub 2015 Aug 15. J Pharmacol Sci. 2015. PMID: 26318672 - [Mechanisms underlying the pathogenesis of neuropathic pain revealing by the role of glial cells].
Tsuda M. Tsuda M. Nihon Shinkei Seishin Yakurigaku Zasshi. 2015 Feb;35(1):1-4. Nihon Shinkei Seishin Yakurigaku Zasshi. 2015. PMID: 25816633 Review. Japanese. - Microglial regulation of neuropathic pain.
Tsuda M, Masuda T, Tozaki-Saitoh H, Inoue K. Tsuda M, et al. J Pharmacol Sci. 2013;121(2):89-94. doi: 10.1254/jphs.12r14cp. Epub 2013 Jan 22. J Pharmacol Sci. 2013. PMID: 23337437 Review.
Cited by
- Deep learning predicts the impact of regulatory variants on cell-type-specific enhancers in the brain.
Zheng A, Shen Z, Glass CK, Gymrek M. Zheng A, et al. Bioinform Adv. 2023 Jan 12;3(1):vbad002. doi: 10.1093/bioadv/vbad002. eCollection 2023. Bioinform Adv. 2023. PMID: 36726730 Free PMC article. - Elevated TNF-α Leads to Neural Circuit Instability in the Absence of Interferon Regulatory Factor 8.
Feinberg PA, Becker SC, Chung L, Ferrari L, Stellwagen D, Anaclet C, Durán-Laforet V, Faust TE, Sumbria RK, Schafer DP. Feinberg PA, et al. J Neurosci. 2022 Aug 10;42(32):6171-6185. doi: 10.1523/JNEUROSCI.0601-22.2022. Epub 2022 Jul 5. J Neurosci. 2022. PMID: 35790400 Free PMC article. - Biallelic interferon regulatory factor 8 mutation: A complex immunodeficiency syndrome with dendritic cell deficiency, monocytopenia, and immune dysregulation.
Bigley V, Maisuria S, Cytlak U, Jardine L, Care MA, Green K, Gunawan M, Milne P, Dickinson R, Wiscombe S, Parry D, Doffinger R, Laurence A, Fonseca C, Stoevesandt O, Gennery A, Cant A, Tooze R, Simpson AJ, Hambleton S, Savic S, Doody G, Collin M. Bigley V, et al. J Allergy Clin Immunol. 2018 Jun;141(6):2234-2248. doi: 10.1016/j.jaci.2017.08.044. Epub 2017 Nov 8. J Allergy Clin Immunol. 2018. PMID: 29128673 Free PMC article. - Purinergic signaling in microglia in the pathogenesis of neuropathic pain.
Inoue K. Inoue K. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(4):174-182. doi: 10.2183/pjab.93.011. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28413195 Free PMC article. Review. - A novel P2X4 receptor-selective antagonist produces anti-allodynic effect in a mouse model of herpetic pain.
Matsumura Y, Yamashita T, Sasaki A, Nakata E, Kohno K, Masuda T, Tozaki-Saitoh H, Imai T, Kuraishi Y, Tsuda M, Inoue K. Matsumura Y, et al. Sci Rep. 2016 Aug 31;6:32461. doi: 10.1038/srep32461. Sci Rep. 2016. PMID: 27576299 Free PMC article.
References
- Burnstock G. Purinergic signalling and disorders of the central nervous system. Nat. Rev. Drug Discov. 2008;7:575–590. - PubMed
- Coull JA, Beggs S, Boudreau D, Boivin D, Tsuda M, Inoue K, Gravel C, Salter MW, De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438:1017–1021. - PubMed
- De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, et al. International MS Genetics Consortium Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials