Clocks, metabolism, and the epigenome - PubMed (original) (raw)
Review
Clocks, metabolism, and the epigenome
Dan Feng et al. Mol Cell. 2012.
Abstract
Many behaviors and physiological activities in living organisms display circadian rhythms, allowing the organisms to anticipate and prepare for the diurnal changes in the living environment. In this way, metabolic processes are aligned with the periodic environmental changes and behavioral cycles, such as the sleep/wake and fasting/feeding cycles. Disturbances of this alignment significantly increase the risk of metabolic diseases. Meanwhile, the circadian clock receives signals from the environment and feedback from metabolic pathways, and adjusts its activity and function. Growing evidence connects the circadian clock with epigenomic regulators. Here we review the recent advances in understanding the crosstalk between the circadian clock and energy metabolism through epigenomic programming and transcriptional regulation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
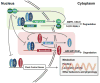
Figure 1. The basic clock machinery consists of negative transcriptional-translational feedback loops
In the first loop, BMAL1/CLOCK drives Per/Cry transcription, while PER/CRY binds and inhibits transcriptional activity of BMAL1/CLOCK. In the second loop, BMAL1/CLOCK drives REV-ERB expression, which in turn represses Bmal1 transcription. Both loops are essential for maintaining circadian rhythm. In addition, post-translational modification, as shown for PER/CRY and REV-ERB, is also important in regulating clock activity. The core clock machinery can drive rhythmic behavioral and physiological activities, such as metabolism.
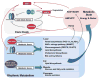
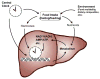
Figure 2. Overview of the interplays between environment, circadian clocks and metabolism
The circadian clock in the cells comprising metabolic organs, such as liver, functions as an epigenomic programmer and controls metabolic outputs. This autonomous apparatus is regulated by the central clock in the SCN and by food intake via hormones and nutrients/metabolites. Both the central clock and food availability contributes to the temporal regulation of food intake.
Figure 3. The core clock machinery drives rhythmic metabolic activities and receives feedback from intracellular metabolites
BMAL1/CLOCK and REV-ERB drive rhythmic metabolic outputs, including NAD+ and heme biosynthesis, while intracellular NAD+ and heme feedback on the clock through their sensors, SIRT1 and REV-ERB, respectively. Intracelluar AMP levels regulates the circadian clock through activation of AMPK and degradation of PER and CRY. The core clock also drives many metabolic pathways in different tissues, which also contribute to the intracellular metabolite pool and metabolic state.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Current Biology. 2002;12:540–550. - PubMed
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. - PubMed
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-Ribose) Polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. - PubMed
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK045586/DK/NIDDK NIH HHS/United States
- RC1 DK086239/DK/NIDDK NIH HHS/United States
- DK43806/DK/NIDDK NIH HHS/United States
- R37 DK043806/DK/NIDDK NIH HHS/United States
- DK45586/DK/NIDDK NIH HHS/United States
- R01 DK043806/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources