Prevention of overfitting in cryo-EM structure determination - PubMed (original) (raw)
- PMID: 22842542
- PMCID: PMC4912033
- DOI: 10.1038/nmeth.2115
Prevention of overfitting in cryo-EM structure determination
Sjors H W Scheres et al. Nat Methods. 2012 Sep.
No abstract available
Figures
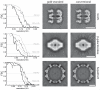
Figure 1. The prevention of overfitting
Tests with three cryo-EM data sets (GroEL, β-galactosidase and hepatitis B) illustrate that overfitting may be avoided without compromising resolution. FSCs between reconstructions from random halves of the data (solid lines) and FSCs between the reconstruction from all particles and the crystal structures (dashed lines) show that the gold-standard procedure (black) does not yield lower resolutions, while the conventional procedure (grey) over-estimates resolution (left column). The frequency where the dashed lines pass through FSC=0.5 indicate the true resolution of the reconstructions from all particles, whereas the frequency where the solid lines pass through FSC=0.143 indicate the reported resolution. Corresponding values are given in Supplementary Table 1. Central slices through the reconstructions for the gold-standard procedure show less noisy maps with fewer artefacts (middle column) than for the conventional procedure (right column). Scale bars (white) correspond to 100 Å.
Similar articles
- Difference mapping cryo-EM.
Peters R, Sikorski R. Peters R, et al. Science. 1999 Feb 19;283(5405):1133. doi: 10.1126/science.283.5405.1133a. Science. 1999. PMID: 10075576 No abstract available. - Fragment-based drug discovery using cryo-EM.
Saur M, Hartshorn MJ, Dong J, Reeks J, Bunkoczi G, Jhoti H, Williams PA. Saur M, et al. Drug Discov Today. 2020 Mar;25(3):485-490. doi: 10.1016/j.drudis.2019.12.006. Epub 2019 Dec 23. Drug Discov Today. 2020. PMID: 31877353 Review. - A Super-Clustering Approach for Fully Automated Single Particle Picking in Cryo-EM.
Al-Azzawi A, Ouadou A, Tanner JJ, Cheng J. Al-Azzawi A, et al. Genes (Basel). 2019 Aug 30;10(9):666. doi: 10.3390/genes10090666. Genes (Basel). 2019. PMID: 31480377 Free PMC article. - Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy.
Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Ludtke SJ, et al. Structure. 2004 Jul;12(7):1129-36. doi: 10.1016/j.str.2004.05.006. Structure. 2004. PMID: 15242589 - The 2017 Nobel Prize in Chemistry: cryo-EM comes of age.
Shen PS. Shen PS. Anal Bioanal Chem. 2018 Mar;410(8):2053-2057. doi: 10.1007/s00216-018-0899-8. Epub 2018 Feb 9. Anal Bioanal Chem. 2018. PMID: 29423601 Review.
Cited by
- The AAA+ ATPase Msp1 is a processive protein translocase with robust unfoldase activity.
Castanzo DT, LaFrance B, Martin A. Castanzo DT, et al. Proc Natl Acad Sci U S A. 2020 Jun 30;117(26):14970-14977. doi: 10.1073/pnas.1920109117. Epub 2020 Jun 15. Proc Natl Acad Sci U S A. 2020. PMID: 32541053 Free PMC article. - Characterization of an Escherichia coli-derived triple-type chimeric vaccine against human papillomavirus types 39, 68 and 70.
Qian C, Yang Y, Xu Q, Wang Z, Chen J, Chi X, Yu M, Gao F, Xu Y, Lu Y, Sun H, Shen J, Wang D, Zhou L, Li T, Wang Y, Zheng Q, Yu H, Zhang J, Gu Y, Xia N, Li S. Qian C, et al. NPJ Vaccines. 2022 Oct 31;7(1):134. doi: 10.1038/s41541-022-00557-y. NPJ Vaccines. 2022. PMID: 36316367 Free PMC article. - Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations.
Liu Y, Zhang Y, Xue H, Cao M, Bai G, Mu Z, Yao Y, Sun S, Fang D, Huang J. Liu Y, et al. Cell Discov. 2021 May 11;7(1):32. doi: 10.1038/s41421-021-00261-6. Cell Discov. 2021. PMID: 33972509 Free PMC article. - SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration.
Figley MD, Gu W, Nanson JD, Shi Y, Sasaki Y, Cunnea K, Malde AK, Jia X, Luo Z, Saikot FK, Mosaiab T, Masic V, Holt S, Hartley-Tassell L, McGuinness HY, Manik MK, Bosanac T, Landsberg MJ, Kerry PS, Mobli M, Hughes RO, Milbrandt J, Kobe B, DiAntonio A, Ve T. Figley MD, et al. Neuron. 2021 Apr 7;109(7):1118-1136.e11. doi: 10.1016/j.neuron.2021.02.009. Epub 2021 Mar 2. Neuron. 2021. PMID: 33657413 Free PMC article. - Structure of Merkel Cell Polyomavirus Capsid and Interaction with Its Glycosaminoglycan Attachment Receptor.
Bayer NJ, Januliene D, Zocher G, Stehle T, Moeller A, Blaum BS. Bayer NJ, et al. J Virol. 2020 Sep 29;94(20):e01664-19. doi: 10.1128/JVI.01664-19. Print 2020 Sep 29. J Virol. 2020. PMID: 32699083 Free PMC article.
References
- Saxton WO, Baumeister W. J. Microscopy. 1982;127:127–138. - PubMed
- Stewart A, Grigorieff N. Ultramicroscopy. 2004;102:67–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources