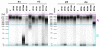Storage conditions of intestinal microbiota matter in metagenomic analysis - PubMed (original) (raw)
Storage conditions of intestinal microbiota matter in metagenomic analysis
Silvia Cardona et al. BMC Microbiol. 2012.
Abstract
Background: The structure and function of human gut microbiota is currently inferred from metagenomic and metatranscriptomic analyses. Recovery of intact DNA and RNA is therefore a critical step in these studies. Here, we evaluated how different storage conditions of fecal samples affect the quality of extracted nucleic acids and the stability of their microbial communities.
Results: We assessed the quality of genomic DNA and total RNA by microcapillary electrophoresis and analyzed the bacterial community structure by pyrosequencing the 16S rRNA gene. DNA and RNA started to fragment when samples were kept at room temperature for more than 24 h. The use of RNAse inhibitors diminished RNA degradation but this protection was not consistent among individuals. DNA and RNA degradation also occurred when frozen samples were defrosted for a short period (1 h) before nucleic acid extraction. The same conditions that affected DNA and RNA integrity also altered the relative abundance of most taxa in the bacterial community analysis. In this case, intra-individual variability of microbial diversity was larger than inter-individual one.
Conclusions: Though this preliminary work explored a very limited number of parameters, the results suggest that storage conditions of fecal samples affect the integrity of DNA and RNA and the composition of their microbial community. For optimal preservation, stool samples should be kept at room temperature and brought at the laboratory within 24 h after collection or be stored immediately at -20°C in a home freezer and transported afterwards in a freezer pack to ensure that they do not defrost at any time. Mixing the samples with RNAse inhibitors outside the laboratory is not recommended since proper homogenization of the stool is difficult to monitor.
Figures
Figure 1
Fragmentation analysis of genomic DNA. Microcapillary electrophoresis patterns of genomic DNA extracted from fecal samples collected by 4 individuals (#1, #2, #3, #4) and stored in the following conditions: immediately frozen at −20°C (F); immediately frozen and then unfrozen during 1 h and 3 h (UF1h, UF3h); kept at room temperature during 3 h, 24 h and 2 weeks (RT3h, RT24h, RT2w). The equivalent to 1 mg of fecal material is loaded on each lane. A DNA fragment size (base pair) ladder was loaded in the left most lanes.
Figure 2
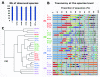
Bacterial community analysis based on 16S rRNA gene survey. A) Alpha-diversity analysis of number of species observed in 6 storage conditions: Immediately frozen (F); unfrozen 1 h and 3 h (UF1h, UF3h); room temperature 3 h, 24 h, and 2 weeks (RT3h, RT24h, RT2w). The plot averages the number of species from the samples provided by 4 individuals in each condition. B) Taxonomy analysis at the species level of the 24 samples based on alignment performed using PyNast against Silva 108 release database and OTUs assignment using blast and the Silva 108 release taxa mapping file. Individual #1 (red), #2 (blue), #3 (green), #4 (purple). A more detailed taxonomy assignment is provided in the additional data (See Additional file 3: Table S1). C) UPGMA clustering of the 24 samples based on weighted UniFrac method. Samples from the 4 individuals are colored as in B. The scale bar represents 2% sequence divergence.
Figure 3
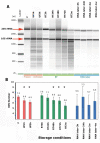
RNA quality analysis. A) Microcapillary electrophoresis patterns of total RNA extracted from fecal samples of one individual that underwent 12 different storage conditions: immediately frozen at −20°C (F); immediately frozen and then unfrozen (UF) during 1 h and 3 h; kept at room temperature (RT) during 3 h, 24 h, 48 h, 72 h and 2 weeks; mixed with commercial RNAse inhibitor solution (RNA later) and kept at room temperature during 3 h, 24 h, 2 weeks and 4 weeks. The equivalent to 1 mg of fecal material is loaded on each lane. A RNA fragment size (nt) marker was loaded in the first lane from the left side. B) Summary plot of average RNA integrity numbers (RIN) obtained with samples stored in the above 12 conditions. N = 11 individuals for the 88 samples stored without RNAse inhibitor. Standard deviation is indicated for each storage condition. N = 6 individuals for the 24 samples stored with RNAse inhibitor. Statistical analysis was performed using Poisson regression model (the star (*) means that the comparison with the frozen sample RIN number was significant with p < 0.05).
Similar articles
- Characterization of the fecal microbiota using high-throughput sequencing reveals a stable microbial community during storage.
Carroll IM, Ringel-Kulka T, Siddle JP, Klaenhammer TR, Ringel Y. Carroll IM, et al. PLoS One. 2012;7(10):e46953. doi: 10.1371/journal.pone.0046953. Epub 2012 Oct 5. PLoS One. 2012. PMID: 23071673 Free PMC article. - Comparison of methods for fecal microbiome biospecimen collection.
Dominianni C, Wu J, Hayes RB, Ahn J. Dominianni C, et al. BMC Microbiol. 2014 Apr 23;14:103. doi: 10.1186/1471-2180-14-103. BMC Microbiol. 2014. PMID: 24758293 Free PMC article. - Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples.
Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Lauber CL, et al. FEMS Microbiol Lett. 2010 Jun;307(1):80-6. doi: 10.1111/j.1574-6968.2010.01965.x. Epub 2010 Mar 25. FEMS Microbiol Lett. 2010. PMID: 20412303 Free PMC article. - Metagenomics of human microbiome: beyond 16s rDNA.
Gosalbes MJ, Abellan JJ, Durbán A, Pérez-Cobas AE, Latorre A, Moya A. Gosalbes MJ, et al. Clin Microbiol Infect. 2012 Jul;18 Suppl 4:47-9. doi: 10.1111/j.1469-0691.2012.03865.x. Clin Microbiol Infect. 2012. PMID: 22647049 Review. - Nucleic acid-based methods to assess the composition and function of the bowel microbiota.
Lawley B, Tannock GW. Lawley B, et al. Gastroenterol Clin North Am. 2012 Dec;41(4):855-68. doi: 10.1016/j.gtc.2012.08.010. Epub 2012 Sep 28. Gastroenterol Clin North Am. 2012. PMID: 23101691 Review.
Cited by
- Effects of Specimen Collection Methodologies and Storage Conditions on the Short-Term Stability of Oral Microbiome Taxonomy.
Luo T, Srinivasan U, Ramadugu K, Shedden KA, Neiswanger K, Trumble E, Li JJ, McNeil DW, Crout RJ, Weyant RJ, Marazita ML, Foxman B. Luo T, et al. Appl Environ Microbiol. 2016 Aug 30;82(18):5519-29. doi: 10.1128/AEM.01132-16. Print 2016 Sep 15. Appl Environ Microbiol. 2016. PMID: 27371581 Free PMC article. - Biomarker development in the precision medicine era: lung cancer as a case study.
Vargas AJ, Harris CC. Vargas AJ, et al. Nat Rev Cancer. 2016 Aug;16(8):525-37. doi: 10.1038/nrc.2016.56. Epub 2016 Jul 8. Nat Rev Cancer. 2016. PMID: 27388699 Free PMC article. Review. - Using fecal immmunochemical cartridges for gut microbiome analysis within a colorectal cancer screening program.
Brezina S, Borkovec M, Baierl A, Bastian F, Futschik A, Gasche N, Gruenberger T, Hallas M, Jannsen C, Leeb G, Lutz R, Sladek B, Gsur A. Brezina S, et al. Gut Microbes. 2023 Jan-Dec;15(1):2176119. doi: 10.1080/19490976.2023.2176119. Gut Microbes. 2023. PMID: 36794815 Free PMC article. - Current State of and Future Opportunities for Prediction in Microbiome Research: Report from the Mid-Atlantic Microbiome Meet-up in Baltimore on 9 January 2019.
Sakowski E, Uritskiy G, Cooper R, Gomes M, McLaren MR, Meisel JS, Mickol RL, Mintz CD, Mongodin EF, Pop M, Rahman MA, Sanchez A, Timp W, Vela JD, Wolz CM, Zackular JP, Chopyk J, Commichaux S, Davis M, Dluzen D, Ganesan SM, Haruna M, Nasko D, Regan MJ, Sarria S, Shah N, Stacy B, Taylor D, DiRuggiero J, Preheim SP. Sakowski E, et al. mSystems. 2019 Oct 8;4(5):e00392-19. doi: 10.1128/mSystems.00392-19. mSystems. 2019. PMID: 31594828 Free PMC article. - The athletic gut microbiota.
Mohr AE, Jäger R, Carpenter KC, Kerksick CM, Purpura M, Townsend JR, West NP, Black K, Gleeson M, Pyne DB, Wells SD, Arent SM, Kreider RB, Campbell BI, Bannock L, Scheiman J, Wissent CJ, Pane M, Kalman DS, Pugh JN, Ortega-Santos CP, Ter Haar JA, Arciero PJ, Antonio J. Mohr AE, et al. J Int Soc Sports Nutr. 2020 May 12;17(1):24. doi: 10.1186/s12970-020-00353-w. J Int Soc Sports Nutr. 2020. PMID: 32398103 Free PMC article. Review.
References
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P. et al.A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S. et al.Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
- Dolfing J, Vos A, Bloem J, Ehlert PA, Naumova NB, Kuikman PJ. Microbial diversity in archived soils. Science. 2004;306(5697):813. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources