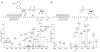Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites - PubMed (original) (raw)
Comment
Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites
Ivan Matic et al. Nat Methods. 2012.
No abstract available
Figures
Figure 1
Fragmentation spectra of apeptide modified by ADP-ribose (a; triply charged precursor at m/z 937.7702) and ribose-phosphate (b; triply charged precursor at m/z 828.0861).
Comment on
- A home for raw proteomics data.
[No authors listed] [No authors listed] Nat Methods. 2012 May;9(5):419. doi: 10.1038/nmeth.2011. Nat Methods. 2012. PMID: 22803195 No abstract available.
Similar articles
- An Advanced Strategy for Comprehensive Profiling of ADP-ribosylation Sites Using Mass Spectrometry-based Proteomics.
Hendriks IA, Larsen SC, Nielsen ML. Hendriks IA, et al. Mol Cell Proteomics. 2019 May;18(5):1010-1026. doi: 10.1074/mcp.TIR119.001315. Epub 2019 Feb 23. Mol Cell Proteomics. 2019. PMID: 30798302 Free PMC article. - The Promise of Proteomics for the Study of ADP-Ribosylation.
Daniels CM, Ong SE, Leung AK. Daniels CM, et al. Mol Cell. 2015 Jun 18;58(6):911-24. doi: 10.1016/j.molcel.2015.06.012. Mol Cell. 2015. PMID: 26091340 Free PMC article. - Reading ADP-ribosylation signaling using chemical biology and interaction proteomics.
Kliza KW, Liu Q, Roosenboom LWM, Jansen PWTC, Filippov DV, Vermeulen M. Kliza KW, et al. Mol Cell. 2021 Nov 4;81(21):4552-4567.e8. doi: 10.1016/j.molcel.2021.08.037. Epub 2021 Sep 21. Mol Cell. 2021. PMID: 34551281 - The phosphoproteomics data explosion.
Lemeer S, Heck AJ. Lemeer S, et al. Curr Opin Chem Biol. 2009 Oct;13(4):414-20. doi: 10.1016/j.cbpa.2009.06.022. Epub 2009 Jul 19. Curr Opin Chem Biol. 2009. PMID: 19620020 Review. - Systems Analysis for Interpretation of Phosphoproteomics Data.
Munk S, Refsgaard JC, Olsen JV. Munk S, et al. Methods Mol Biol. 2016;1355:341-60. doi: 10.1007/978-1-4939-3049-4_23. Methods Mol Biol. 2016. PMID: 26584937 Review.
Cited by
- Site-specific analysis of the Asp- and Glu-ADP-ribosylated proteome by quantitative mass spectrometry.
Li P, Zhen Y, Yu Y. Li P, et al. Methods Enzymol. 2019;626:301-321. doi: 10.1016/bs.mie.2019.06.024. Epub 2019 Jul 24. Methods Enzymol. 2019. PMID: 31606080 Free PMC article. - ADP-ribosylation signalling and human disease.
Palazzo L, Mikolčević P, Mikoč A, Ahel I. Palazzo L, et al. Open Biol. 2019 Apr 26;9(4):190041. doi: 10.1098/rsob.190041. Open Biol. 2019. PMID: 30991935 Free PMC article. Review. - Serine ADP-Ribosylation Depends on HPF1.
Bonfiglio JJ, Fontana P, Zhang Q, Colby T, Gibbs-Seymour I, Atanassov I, Bartlett E, Zaja R, Ahel I, Matic I. Bonfiglio JJ, et al. Mol Cell. 2017 Mar 2;65(5):932-940.e6. doi: 10.1016/j.molcel.2017.01.003. Epub 2017 Feb 9. Mol Cell. 2017. PMID: 28190768 Free PMC article. - 2016 update of the PRIDE database and its related tools.
Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. Vizcaíno JA, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D447-56. doi: 10.1093/nar/gkv1145. Epub 2015 Nov 2. Nucleic Acids Res. 2016. PMID: 26527722 Free PMC article. - Characterization of TCDD-inducible poly-ADP-ribose polymerase (TIPARP/ARTD14) catalytic activity.
Gomez A, Bindesbøll C, Satheesh SV, Grimaldi G, Hutin D, MacPherson L, Ahmed S, Tamblyn L, Cho T, Nebb HI, Moen A, Anonsen JH, Grant DM, Matthews J. Gomez A, et al. Biochem J. 2018 Dec 11;475(23):3827-3846. doi: 10.1042/BCJ20180347. Biochem J. 2018. PMID: 30373764 Free PMC article.
References
- Nat. Methods. 2012;9:419–419. - PubMed
- Laing S, Koch-Nolte F, Haag F, Buck F. J. Proteomics. 2011;75:169–176. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources