Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn's disease - PubMed (original) (raw)
Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn's disease
Melanie Craven et al. PLoS One. 2012.
Abstract
Background and aims: Understanding the interplay between genetic susceptibility, the microbiome, the environment and the immune system in Crohn's Disease (CD) is essential for developing optimal therapeutic strategies. We sought to examine the dynamics of the relationship between inflammation, the ileal microbiome, and host genetics in murine models of ileitis.
Methods: We induced ileal inflammation of graded severity in C57BL6 mice by gavage with Toxoplasma gondii, Giardia muris, low dose indomethacin (LDI; 0.1 mg/mouse), or high dose indomethacin (HDI; 1 mg/mouse). The composition and spatial distribution of the mucosal microbiome was evaluated by 16S rDNA pyrosequencing and fluorescence in situ hybridization. Mucosal E. coli were enumerated by quantitative PCR, and characterized by phylogroup, genotype and pathotype.
Results: Moderate to severe ileitis induced by T. gondii (day 8) and HDI caused a consistent shift from >95% gram + Firmicutes to >95% gram - Proteobacteria. This was accompanied by reduced microbial diversity and mucosal invasion by adherent and invasive E. coli, mirroring the dysbiosis of ileal CD. In contrast, dysbiosis and bacterial invasion did not develop in mice with mild ileitis induced by Giardia muris. Superimposition of genetic susceptibility and T. Gondii infection revealed greatest dysbiosis and bacterial invasion in the CD-susceptible genotype, NOD2(-/-), and reduced dysbiosis in ileitis-resistant CCR2(-/-) mice. Abrogating inflammation with the CD therapeutic anti-TNF-α-mAb tempered dysbiosis and bacterial invasion.
Conclusions: Acute ileitis induces dysbiosis and proliferation of mucosally invasive E. coli, irrespective of trigger and genotype. The identification of CCR2 as a target for therapeutic intervention, and discovery that host genotype and therapeutic blockade of inflammation impact the threshold and extent of ileal dysbiosis are of high relevance to developing effective therapies for CD.
Conflict of interest statement
Competing Interests: Dr. Scot Dowd is an employee of MR DNA (Molecular Research), Shallowater, TX, and performed the 16S pyrosequencing analysis. This does not represent a conflict of interest. The pyrosequencing was performed in a blinded fashion. Dr. Dowd is an expert in the analysis of 16S data and helped to evaluate the consequences of intestinal inflammation on the intestinal microbiome. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Figures
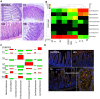
Figure 1. T. gondii infection and high dose indomethacin trigger severe ileitis, dysbiosis and mucosal E. coli invasion in C57BL/6 mice.
(a) Histopathology. Normal ileal histology (inflammation score median = 0, range 0–0) in control mice. Moderate to severe ileitis (4, 3–6) develops in T. gondii mice at 8 days post-infection (T8). _G. muris_-infected mice develop minimal inflammation (1, 1–1) and mucosal hyperplasia 14 days (G14). High dose indomethacin (HDI) induces severe ileitis (5, 5–7) (H&E 40x). (b) Pyrosequencing reveals a Gram− shift associated with ileitis. Gram+ bacteria predominate in control mice and mice with mild inflammation (G7, G14: Giardia 7 and 14 days p.i., T4: T. gondii 4 days p.i.; LDI: low-dose indomethacin). A shift to >95% Gram− bacteria dominated by the phyla Proteobacteria and Bacteroidetes (p<0.0001) is associated with moderate to severe ileitis in T8 and HDI mice. (c) Association plots of the number of sequences obtained by pyrosequencing corresponding to bacterial families. The area of each rectangle is proportional to the difference in observed and expected frequencies on chi square analysis. The observed frequency of a rectangle is indicated by position relative to baseline, increased (shaded red) or decreased (shaded green) highlighting the ileitis-associated shifts from Firmicutes (Lactobacillaceae, Clostridiales) to Enterobacteriaceae (T8, HDI) and Erysipelotrichaceae (G7, G14, T4). (d) Eubacterial FISH reveals scant luminal and mucosa-associated flora in control mice (EUB338-Cy3, non-EUB338-6FAM with DAPI counterstaining, 40x). Ileitis in T8 and HDI is associated with increased mucosal bacteria and invasive E. coli (E. _coli_-Cy3, EUB338-6FAM, 40x).
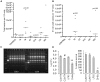
Figure 2. Ileitis-induced dysbiosis is associated with increased bacteria and clonal proliferation of adherent invasive E. coli (AIEC).
(a) Total bacteria quantified by real-time PCR, expressed as bacterial CFU per 106 murine cells. Moderate to severe ileitis in T8 is associated with increased bacteria relative to controls. Mild pathology (T4, G14) induces a decrease. (b) Moderate to severe ileitis (T8, HDI) induces E. coli proliferation (E. coli uidA quantification by real-time PCR). (c) Agarose gel electrophoresis showing the products of Random Amplification of Polymorphic DNA (RAPD-PCR) using primers 1283 and 1254. E. coli strains from: T4 (Lanes 1–3), T8 (4, 5) and HDI (6–8). Lane L, 100bp plus DNA ladder. E. coli isolates in lanes 2–8 are clonal, and a representative strain was designated CUMT8. E. coli in lane 1 was designated CUMT4. (d). Invasion and survival of E. coli CUMT8 and CUMT4 in cultured epithelial cells (Caco-2) and macrophages (J774). CUMT8 and CUMT4 invade, and persist intracellularly like CD-associated AIEC LF82, and better than commensal E. coli DH5α (* = P<0.01, ** = P<0.001 vs other strains). CUMT4 was less invasive than CUMT8 (# = P<0.01).
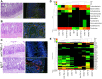
Figure 3. Dysbiosis and E. coli invasion are modulated by genetic susceptibility and pharmacotherapy.
(a) CCR2−/−8 (n = 5) are protected from _T. gondii-_induced ileitis (median inflammation score 3, range 2–3) and E. coli invasion. (b) NOD2−/−8 (n = 5) develop severe ileitis (5, 5–7) and E. coli invasion. (c) anti-TNF-α mAb-7 (n = 5) reduces ileitis (4, 2–4 vs 4, 4–7) and decreases bacterial invasion relative to IgG-7 controls, n = 5). Histology, H&E 40x; FISH, CCR2−/−8 and anti-TNF-α-7 = Cy3-EUB338/non-EUB338-6FAM; NOD2−/−8 and IgG-7 = Cy3-E. coli/EUB338-6FAM, 40x. (d,e). Pyrosequencing reveals that _T. gondii_-induced dysbiosis is modulated by genetic susceptibility and pharmacotherapy. Abrogating ileitis maintains microbial diversity (CCR2−/−8), whereas enhancing ileitis decreases diversity and increases E. coli (NOD2−/−8). NOD2 deletion is associated with a baseline shift to Bacteroidetes (NOD2−/−0). Anti-TNF-α mAb temper dysbiosis (anti-TNF-α-7 versus IgG-7).
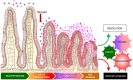
Figure 4. Inflammation drives dysbiosis, Gram negative proliferation and E. coli invasion.
Independent of genotype, ileitis induces a progressive decrease in microbial diversity, a shift from Firmicutes (Gram**+**) to Proteobacteria (Gram−), and proliferation of AIEC. Superimposition of genetic susceptibility can lower (NOD2−/−) or increase (CCR2−/−) the threshold for dysbiosis in response to an external trigger. We speculate that genetic susceptibility may also influence the ability of an individual to resolve the self-perpetuating cycle of dysbiosis and inflammation generated by an acute insult.
Similar articles
- Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation.
Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, Stewart K, Scherl EJ, Araz Y, Bitar PP, Lefébure T, Chandler B, Schukken YH, Stanhope MJ, Simpson KW. Dogan B, et al. Inflamm Bowel Dis. 2014 Nov;20(11):1919-32. doi: 10.1097/MIB.0000000000000183. Inflamm Bowel Dis. 2014. PMID: 25230163 - IL23R-Protective Coding Variant Promotes Beneficial Bacteria and Diversity in the Ileal Microbiome in Healthy Individuals Without Inflammatory Bowel Disease.
Zakrzewski M, Simms LA, Brown A, Appleyard M, Irwin J, Waddell N, Radford-Smith GL. Zakrzewski M, et al. J Crohns Colitis. 2019 Mar 30;13(4):451-461. doi: 10.1093/ecco-jcc/jjy188. J Crohns Colitis. 2019. PMID: 30445599 - Genetic deletion of the bacterial sensor NOD2 improves murine Crohn's disease-like ileitis independent of functional dysbiosis.
Corridoni D, Rodriguez-Palacios A, Di Stefano G, Di Martino L, Antonopoulos DA, Chang EB, Arseneau KO, Pizarro TT, Cominelli F. Corridoni D, et al. Mucosal Immunol. 2017 Jul;10(4):971-982. doi: 10.1038/mi.2016.98. Epub 2016 Nov 16. Mucosal Immunol. 2017. PMID: 27848951 Free PMC article. - Postsurgical recurrence of ileal Crohn's disease: an update on risk factors and intervention points to a central role for impaired host-microflora homeostasis.
Cunningham MF, Docherty NG, Coffey JC, Burke JP, O'Connell PR. Cunningham MF, et al. World J Surg. 2010 Jul;34(7):1615-26. doi: 10.1007/s00268-010-0504-6. World J Surg. 2010. PMID: 20195604 Review. - Insights into inflammatory bowel disease using Toxoplasma gondii as an infectious trigger.
Egan CE, Cohen SB, Denkers EY. Egan CE, et al. Immunol Cell Biol. 2012 Aug;90(7):668-75. doi: 10.1038/icb.2011.93. Epub 2011 Nov 8. Immunol Cell Biol. 2012. PMID: 22064707 Free PMC article. Review.
Cited by
- Gastroenterology issues in schizophrenia: why the gut matters.
Severance EG, Prandovszky E, Castiglione J, Yolken RH. Severance EG, et al. Curr Psychiatry Rep. 2015 May;17(5):27. doi: 10.1007/s11920-015-0574-0. Curr Psychiatry Rep. 2015. PMID: 25773227 Free PMC article. Review. - Depletion of dietary aryl hydrocarbon receptor ligands alters microbiota composition and function.
Brawner KM, Yeramilli VA, Duck LW, Van Der Pol W, Smythies LE, Morrow CD, Elson CO, Martin CA. Brawner KM, et al. Sci Rep. 2019 Oct 11;9(1):14724. doi: 10.1038/s41598-019-51194-w. Sci Rep. 2019. PMID: 31604984 Free PMC article. - Effects of the Lipid Profile, Type 2 Diabetes and Medication on the Metabolic Syndrome-Associated Gut Microbiome.
Gradisteanu Pircalabioru G, Liaw J, Gundogdu O, Corcionivoschi N, Ilie I, Oprea L, Musat M, Chifiriuc MC. Gradisteanu Pircalabioru G, et al. Int J Mol Sci. 2022 Jul 6;23(14):7509. doi: 10.3390/ijms23147509. Int J Mol Sci. 2022. PMID: 35886861 Free PMC article. - Depletion of Lipocalin 2 (LCN2) in Mice Leads to Dysbiosis and Persistent Colonization with Segmented Filamentous Bacteria.
Klüber P, Meurer SK, Lambertz J, Schwarz R, Zechel-Gran S, Braunschweig T, Hurka S, Domann E, Weiskirchen R. Klüber P, et al. Int J Mol Sci. 2021 Dec 5;22(23):13156. doi: 10.3390/ijms222313156. Int J Mol Sci. 2021. PMID: 34884961 Free PMC article. - Autoimmune phenotypes in schizophrenia reveal novel treatment targets.
Severance EG, Dickerson FB, Yolken RH. Severance EG, et al. Pharmacol Ther. 2018 Sep;189:184-198. doi: 10.1016/j.pharmthera.2018.05.005. Epub 2018 May 6. Pharmacol Ther. 2018. PMID: 29742478 Free PMC article. Review.
References
- Hanauer SB. Inflammatory bowel disease: Epidemiology, pathogenesis, and therapeutic opportunities. Inflamm Bowel Dis. 2006;12:S3–9. - PubMed
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. - PubMed
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. - PubMed
- Singh S, Graff LA, Bernstein CN. Do NSAIDs, antibiotics, infections, or stress trigger flares in IBD? Am J Gastroenterol 104(5): 1298–313; quiz 1314. 2009. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases