Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection - PubMed (original) (raw)
Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection
S Eréndira Avendaño-Vázquez et al. Genes Dev. 2012.
Abstract
TDP-43 is a critical RNA-binding factor associated with pre-mRNA splicing in mammals. Its expression is tightly autoregulated, with loss of this regulation implicated in human neuropathology. We demonstrate that TDP-43 overexpression in humans and mice activates a 3' untranslated region (UTR) intron, resulting in excision of the proximal polyA site (PAS) pA(1). This activates a cryptic PAS that prevents TDP-43 expression through a nuclear retention mechanism. Superimposed on this process, overexpression of TDP-43 blocks recognition of pA(1) by competing with CstF-64 for PAS binding. Overall, we uncover complex interplay between transcription, splicing, and 3' end processing to effect autoregulation of TDP-43.
Figures
Figure 1.
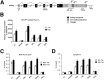
_Cis_-acting elements and molecular events in TDP-43 autoregulation. (A) Schematic diagram of TDP-43 illustrating locations of the stop codon (tag), PASs (pA1–4), the TDPBR, and splicing events (in coding sequences, filled lines; in 3′ UTR, dotted lines). Coding regions (black boxes), untranslated sequences (gray boxes), and introns (connecting black lines) are indicated. RT–PCR primers are indicated. (B, left) Schematic representation of the GFP-3′UTR reporters are shown. (Middle) Western blots of GFP reporter protein in normal (−Tet) and TDP-43 overexpression conditions (+Tet) are shown. (Right) The bar chart shows densitometric quantification of protein expression, normalized against DiGFP expression. Mean values are plotted; error bars indicate the SD from at least three independent experiments. (C) 3′ RACE analysis (−Tet and +Tet) for x7 (left) and x7 Δgt-ag (right) reporters using the specific GFPfw and anchor primers (see A). (Right) The schematic representation denotes amplified mRNA isoforms (bands 1 and 2) confirmed by sequence analysis. (D) 3′ RACE analysis of endogenous TDP-43 (−Tet and +Tet). The forward primer annealed to intron 6 (a), while the anchor was used as the reverse primer (see A). (Right) Amplified mRNA isoforms are indicated.
Figure 2.
Endogenous TDP-43 in mice and humans; mRNA intron 7 splicing, PAS usage, and mRNAs nuclear retention. (A) Schematic representation of human TDP-43. Arrows show the positions of the primers used. 3′ RACE analysis of endogenous TDP-43 before (−Tet) and following (+Tet) TDP-43 transgene induction performed on nuclear (N) and cytoplasmic (C) RNA fractions or following CHX treatment. The forward primer was annealed to intron 6 (a), while the anchor was used as the reverse primer. (Right) The same samples were checked for the amplification of the endogenous variant V2 mRNA using primers b and c as indicated. All PCR products were sequenced to establish their identity. Distribution of p84 and tubulin was analyzed by Western blotting. (B, left) Schematic diagram of mouse TDP-43 showing sequence conservation between humans and mice and detailed alignment of the sequence corresponding to intron 7 splice sites and the various PAS. (Right) 3′ RACE analysis of endogenous murine TDP-43 from control (wild-type [wt]) and transgenic tg A315T mouse brains. The forward primer is annealed on intron 6 (named ma), while the anchor was used as the reverse primer. A representation of amplified mRNA isoforms (bands 1 and 2) is also shown as confirmed by sequencing. HPRT mRNA amplification was used as a control. (Bottom right) Endogenous and transgenic tg A315T protein expression was measured by Western blotting using anti-TDP-43, with anti-tubulin as control.
Figure 3.
Pol II stalling on the TDPBR sequence. (A) Schematic diagram of TDP-43 showing positions of ChIP probes used. (B) BrU nuclear run-on assay measuring nascent transcription in different regions of endogenous TDP-43 under normal conditions (−Tet) and following TDP-43 overexpression (+Tet). (C,D) ChIP analysis using N20 Pol II and H5 Pol II CTDser2P antibodies probing the same regions of TDP-43.
Figure 4.
Competition between TDP-43 and CstF-64 across the pA1 site. (A) Nucleotide sequence of TDP-43 pA1 DSE (top) and a hit frequency plot of TDP-43 CLIP data (bottom) (from the University of California at Santa Cruz Genome Browser). (B) RIP analysis of TDP-43 and CstF-64 proteins on endogenous TDP-43 across pA1 under normal (−Tet) and TDP-43 overexpression (+Tet) conditions. (C) RIP for TDP-43 and CstF-64 proteins across the HPRT PAS used as control.
Figure 5.
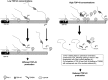
Model of TDP-43 autoregulatory mechanism. Under normal conditions, Pol II synthesizes the nascent TDP-43 mRNA from TDP-43, generating two main TDP-43 mRNA isoforms using either pA1 or pA4, which results in efficient TDP-43 protein expression. When the levels of nuclear TDP-43 rise, there is an increase in the binding of TDP-43 to the TDPBR. This promotes a dual effect: increased intron 7 splicing with the physical removal of the pA1 signal, and direct competition with the cleavage polyA factor Cstf-64 over pA1. Moreover, Pol II stalling may lead to premature termination of transcription and rapid degradation.
Similar articles
- Predominance of spliceosomal complex formation over polyadenylation site selection in TDP-43 autoregulation.
Bembich S, Herzog JS, De Conti L, Stuani C, Avendaño-Vázquez SE, Buratti E, Baralle M, Baralle FE. Bembich S, et al. Nucleic Acids Res. 2014 Mar;42(5):3362-71. doi: 10.1093/nar/gkt1343. Epub 2013 Dec 24. Nucleic Acids Res. 2014. PMID: 24369426 Free PMC article. - An intron enhancer recognized by splicing factors activates polyadenylation.
Lou H, Gagel RF, Berget SM. Lou H, et al. Genes Dev. 1996 Jan 15;10(2):208-19. doi: 10.1101/gad.10.2.208. Genes Dev. 1996. PMID: 8566754 - The polyA tail facilitates splicing of last introns with weak 3' splice sites via PABPN1.
Huang L, Li G, Du C, Jia Y, Yang J, Fan W, Xu YZ, Cheng H, Zhou Y. Huang L, et al. EMBO Rep. 2023 Oct 9;24(10):e57128. doi: 10.15252/embr.202357128. Epub 2023 Sep 4. EMBO Rep. 2023. PMID: 37661812 Free PMC article. - The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5' capping and splicing.
Yang Q, Gilmartin GM, Doublié S. Yang Q, et al. RNA Biol. 2011 Sep-Oct;8(5):748-53. doi: 10.4161/rna.8.5.16040. Epub 2011 Sep 1. RNA Biol. 2011. PMID: 21881408 Free PMC article. Review. - Implications of polyadenylation in health and disease.
Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Curinha A, et al. Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
- Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions.
Nussbacher JK, Tabet R, Yeo GW, Lagier-Tourenne C. Nussbacher JK, et al. Neuron. 2019 Apr 17;102(2):294-320. doi: 10.1016/j.neuron.2019.03.014. Neuron. 2019. PMID: 30998900 Free PMC article. Review. - Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis.
Yang C, Wang H, Qiao T, Yang B, Aliaga L, Qiu L, Tan W, Salameh J, McKenna-Yasek DM, Smith T, Peng L, Moore MJ, Brown RH Jr, Cai H, Xu Z. Yang C, et al. Proc Natl Acad Sci U S A. 2014 Mar 25;111(12):E1121-9. doi: 10.1073/pnas.1322641111. Epub 2014 Mar 10. Proc Natl Acad Sci U S A. 2014. PMID: 24616503 Free PMC article. - Loss of ALS-associated TDP-43 in zebrafish causes muscle degeneration, vascular dysfunction, and reduced motor neuron axon outgrowth.
Schmid B, Hruscha A, Hogl S, Banzhaf-Strathmann J, Strecker K, van der Zee J, Teucke M, Eimer S, Hegermann J, Kittelmann M, Kremmer E, Cruts M, Solchenberger B, Hasenkamp L, van Bebber F, Van Broeckhoven C, Edbauer D, Lichtenthaler SF, Haass C. Schmid B, et al. Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):4986-91. doi: 10.1073/pnas.1218311110. Epub 2013 Mar 1. Proc Natl Acad Sci U S A. 2013. PMID: 23457265 Free PMC article. - TDP-43/FUS in motor neuron disease: Complexity and challenges.
Guerrero EN, Wang H, Mitra J, Hegde PM, Stowell SE, Liachko NF, Kraemer BC, Garruto RM, Rao KS, Hegde ML. Guerrero EN, et al. Prog Neurobiol. 2016 Oct-Nov;145-146:78-97. doi: 10.1016/j.pneurobio.2016.09.004. Epub 2016 Sep 28. Prog Neurobiol. 2016. PMID: 27693252 Free PMC article. Review. - RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia.
McMillan M, Gomez N, Hsieh C, Bekier M, Li X, Miguez R, Tank EMH, Barmada SJ. McMillan M, et al. Mol Cell. 2023 Jan 19;83(2):219-236.e7. doi: 10.1016/j.molcel.2022.12.019. Epub 2023 Jan 11. Mol Cell. 2023. PMID: 36634675 Free PMC article.
References
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE 2008. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 121: 3778–3785 - PubMed
- Buratti E, Baralle FE 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 - PubMed
- Buratti E, Baralle FE 2011. TDP-43: New aspects of autoregulation mechanisms in RNA binding proteins and their connection with human disease. FEBS J 278: 3530–3538 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases