Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630 - PubMed (original) (raw)
Comparative Study
Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630
Nigel G Ternan et al. PLoS One. 2012.
Abstract
Clostridium difficile is considered to be one of the most important causes of health care-associated infections worldwide. In order to understand more fully the adaptive response of the organism to stressful conditions, we examined transcriptional changes resulting from a clinically relevant heat stress (41 °C versus 37 °C) in C. difficile strain 630 and identified 341 differentially expressed genes encompassing multiple cellular functional categories. While the transcriptome was relatively resilient to the applied heat stress, we noted upregulation of classical heat shock genes including the groEL and dnaK operons in addition to other stress-responsive genes. Interestingly, the flagellin gene (fliC) was downregulated, yet genes encoding the cell-wall associated flagellar components were upregulated suggesting that while motility may be reduced, adherence--to mucus or epithelial cells--could be enhanced during infection. We also observed that a number of phage associated genes were downregulated, as were genes associated with the conjugative transposon Tn5397 including a group II intron, thus highlighting a potential decrease in retromobility during heat stress. These data suggest that maintenance of lysogeny and genome wide stabilisation of mobile elements could be a global response to heat stress in this pathogen.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
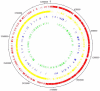
Figure 1. Projection of differentially expressed genes on the Clostridium difficile 630 genome.
Those genes showing at least 1.5-fold change with p<0.05 were selected. From outside to inside: Ring 1, molecular clock of C. difficile strain 630 genome; Ring 2 (red), coding DNA sequences of the forward strand of the genome; Ring 3 (yellow), coding DNA sequences(CDS) of the opposite strand of the genome; Ring 4 (blue), genes upregulated at 41°C; Ring 5 (green), genes downregulated at 41°C.
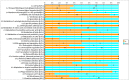
Figure 2. Functional category annotation of differentially expressed genes (>1.5 fold, p<0.05) in the Clostridium difficile strain 630 heat stress transcriptome.
Key: Orange – up regulated; blue – down regulated.
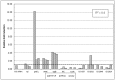
Figure 3. Comparison of qRT-PCR, iTRAQ proteomics and microarray data for selected Clostridium difficile strain 630 genes.
For each individual gene, expressional fold-change values (up-hatched columns) are shown relative to the 37°C control. Corresponding iTRAQ fold-changes (gray columns) are included for comparison with microarray data (down-hatched columns) and show good correlation between the three data sets. 16S rRNA, tpi, and CD2849, whose expression did not change by more than 1.5-fold, were used as reference genes.
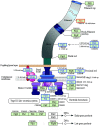
Figure 4. The flagellar motor assembly in Clostridium difficile strain 630.
Graphic is adapted from Kyoto Encyclopedia of Genes and Genomes (
). Several genes involved in the flagellar assembly showed altered expression levels in response to heat-stress. Additional genes found in C. difficile (FlgN, fliA, FliT) were added following BLAST searching. There appear to be no homologues of the master regulator, FlhD/C in C. difficile strain 630. Key: Red box, down regulated >1.5 fold; blue box, up regulated >1.5 fold, black box, no statistically significant change.
Figure 5. Genomic context of Transposon Tn5397 in the genome of Clostridium difficile strain 630.
Key: Arrows represent direction of changes in gene expression; genes lacking an arrow are unchanged. Tn5397 ORF numbers are shown above the genes. CD0506: group II intron. tetM: tetracycline resistance determinant.
Similar articles
- Demonstration that the group II intron from the Clostridial Conjugative transposon Tn5397 undergoes splicing In vivo.
Roberts AP, Braun V, von Eichel-Streiber C, Mullany P. Roberts AP, et al. J Bacteriol. 2001 Feb;183(4):1296-9. doi: 10.1128/JB.183.4.1296-1299.2001. J Bacteriol. 2001. PMID: 11157942 Free PMC article. - Semiquantitative analysis of clinical heat stress in Clostridium difficile strain 630 using a GeLC/MS workflow with emPAI quantitation.
Ternan NG, Jain S, Graham RL, McMullan G. Ternan NG, et al. PLoS One. 2014 Feb 24;9(2):e88960. doi: 10.1371/journal.pone.0088960. eCollection 2014. PLoS One. 2014. PMID: 24586458 Free PMC article. - Quantitative proteomic analysis of the heat stress response in Clostridium difficile strain 630.
Jain S, Graham C, Graham RL, McMullan G, Ternan NG. Jain S, et al. J Proteome Res. 2011 Sep 2;10(9):3880-90. doi: 10.1021/pr200327t. Epub 2011 Aug 8. J Proteome Res. 2011. PMID: 21786815 - Mobile genetic elements in Clostridium difficile and their role in genome function.
Mullany P, Allan E, Roberts AP. Mullany P, et al. Res Microbiol. 2015 May;166(4):361-7. doi: 10.1016/j.resmic.2014.12.005. Epub 2015 Jan 7. Res Microbiol. 2015. PMID: 25576774 Free PMC article. Review. - Expression of heat shock genes in Clostridium acetobutylicum.
Bahl H, Müller H, Behrens S, Joseph H, Narberhaus F. Bahl H, et al. FEMS Microbiol Rev. 1995 Oct;17(3):341-8. doi: 10.1111/j.1574-6976.1995.tb00217.x. FEMS Microbiol Rev. 1995. PMID: 7576772 Review.
Cited by
- The complete catalog of antimicrobial resistance secondary active transporters in Clostridioides difficile: evolution and drug resistance perspective.
Chanket W, Pipatthana M, Sangphukieo A, Harnvoravongchai P, Chankhamhaengdecha S, Janvilisri T, Phanchana M. Chanket W, et al. Comput Struct Biotechnol J. 2024 May 21;23:2358-2374. doi: 10.1016/j.csbj.2024.05.027. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38873647 Free PMC article. - Clostridioides difficile Flagella.
Marvaud JC, Bouttier S, Saunier J, Kansau I. Marvaud JC, et al. Int J Mol Sci. 2024 Feb 12;25(4):2202. doi: 10.3390/ijms25042202. Int J Mol Sci. 2024. PMID: 38396876 Free PMC article. Review. - Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence.
Taggart MG, Snelling WJ, Naughton PJ, La Ragione RM, Dooley JSG, Ternan NG. Taggart MG, et al. PLoS Pathog. 2021 Sep 9;17(9):e1009817. doi: 10.1371/journal.ppat.1009817. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499698 Free PMC article. Review. - What's a Biofilm?-How the Choice of the Biofilm Model Impacts the Protein Inventory of Clostridioides difficile.
Brauer M, Lassek C, Hinze C, Hoyer J, Becher D, Jahn D, Sievers S, Riedel K. Brauer M, et al. Front Microbiol. 2021 Jun 10;12:682111. doi: 10.3389/fmicb.2021.682111. eCollection 2021. Front Microbiol. 2021. PMID: 34177868 Free PMC article. - RidA Proteins Protect against Metabolic Damage by Reactive Intermediates.
Irons JL, Hodge-Hanson K, Downs DM. Irons JL, et al. Microbiol Mol Biol Rev. 2020 Jul 15;84(3):e00024-20. doi: 10.1128/MMBR.00024-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32669283 Free PMC article. Review.
References
- Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature Rev Microbiology 7: 526–536. - PubMed
- Stabler RA, He M, Dawson L, Martin M, Valiente E, et al. (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. BMC Genome Biol 10: R102 doi:10.1186/gb-2009-10-9-r102. - DOI - PMC - PubMed
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, et al. (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467: 711–713. - PubMed
- Bartlett JG (2006) Narrative review: The new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med 145: 758–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
SJ was supported by a Vice Chancellor's Research Scholarship award (2007–2010) from the University of Ulster. GMcM was supported by an Innovation Leaders award (2009) from the HSC Research and Development office (http://www.publichealth.hscni.net/directorate-public-health/hsc-research-and-development). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials