Going viral: next-generation sequencing applied to phage populations in the human gut - PubMed (original) (raw)
Review
Going viral: next-generation sequencing applied to phage populations in the human gut
Alejandro Reyes et al. Nat Rev Microbiol. 2012 Sep.
Abstract
Over the past decade, researchers have begun to characterize viral diversity using metagenomic methods. These studies have shown that viruses, the majority of which infect bacteria, are probably the most genetically diverse components of the biosphere. Here, we briefly review the incipient rise of a phage biology renaissance, which has been catalysed by advances in next-generation sequencing. We explore how work characterizing phage diversity and lifestyles in the human gut is changing our view of ourselves as supra-organisms. Finally, we discuss how a renewed appreciation of phage dynamics may yield new applications for phage therapies designed to manipulate the structure and functions of our gut microbiomes.
Figures
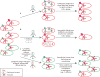
Figure 1. Experimental and computational methods for characterization of phage populations present in the human gut microbiota
See text for further discussion
Figure 2. Potential consequences of a temperate phage lifecycle in the human gut
Viral metagenomic show that the phage population associated with the adult human gut microbiota is characterized by a relatively low number of virotypes compared to other ecosystems (e.g., soils, sediments, marine environments). These gut populations also exhibit high temporal stability of virotypes with respect to both viral community structure and nucleotide sequence conservation, and a high prevalence of temperate phages. These characteristics suggest that a temperate lifestyle is dominant in the distal human gut versus the lytic lifestyle observed in open oceans. (a–c) Illustration of the benefits of this temperate lifestyle on phage-host dynamics. (a) Integration as a prophage protects the host from superinfection, effectively ‘immunizing’ the bacterial host against infection from the same or closely related phages. Furthermore, the genes encoded by the viral genome may expand the niche of the bacterial host by enabling metabolism of new nutrient sources (e.g., carbohydrates), providing antibiotic resistance, conveying virulence factors, or altering host gene expression. This temperate lifecycle allows viral expansion in a 1:1 ratio with the bacterial host. If the integrated virus conveys increased fitness to its bacterial host, there will be increased prevalence of the host and phage in the microbiota. (b) Induction of a lytic cycle may follow a lysogenic state and can be triggered by environmental stress. As a consequence, bacterial turnover is accelerated and energy utilization optimized through ‘phage shunts’, where the debris remaining after lysis is used as a nutrient source by the surviving population. Furthermore, a subpopulation of bacteria that undergoes lytic induction sweeps away other sensitive species and increases the niche for survivors (i.e., bacteria that already have an integrated phage). Periodic induction of prophages can also lead to a constant diversity dynamic , which helps maintain community structure and functional efficiency. (c) Novel infections or infections of novel bacterial hosts by phages bring the benefit of horizontally transferred genes, and create selective pressure on the hosts for diversification of their phage receptors, which are often involved in carbohydrate utilization.
Figure 3. Potential strategies for phage therapy
(a) The traditional strategy has been to use a lytic phage against pathogenic bacteria. While transiently useful, this approach can lead to rapid resistance given positive selection for subpopulations (‘clones’) that are resistant to the lytic phage. Note that this is an antiquated approach to phage therapy that can be trivially improved by using multiple phages with non-overlapping host resistance patterns, or by selecting for phage mutants that overcome host resistance. (b) More recently, synergistic relationships between phage and antibiotics have been exploited, where lysogenic phages are introduced that alone do not kill the pathogen, but instead decrease its survival when used in concert with antibiotics. An example is a phage that inhibits a DNA damage repair system (SOS), which makes bacteria exquisitely sensitive to quinolone-class antibiotics . (c) With our growing understanding of the human microbiome, it may be possible to take a more nuanced approach - selectively manipulating (enhancing) microbial community functions or clearing the way for invasion by probiotic consortia. Strategies can be envisioned to benefit both microbes and their host; for example, introducing genes into phage genomes that are involved in nutrient biosynthesis (with direct benefits to the bacterial and potentially human host), or degradation of nutrients (which may stabilize the representation and niches of beneficial microbes, especially during times of acute stress).
Similar articles
- Does over a century of aerobic phage work provide a solid framework for the study of phages in the gut?
Shareefdeen H, Hynes AP. Shareefdeen H, et al. Anaerobe. 2021 Apr;68:102319. doi: 10.1016/j.anaerobe.2021.102319. Epub 2021 Jan 16. Anaerobe. 2021. PMID: 33465423 Review. - The relationship between the phageome and human health: are bacteriophages beneficial or harmful microbes?
Fernández L, Duarte AC, Rodríguez A, García P. Fernández L, et al. Benef Microbes. 2021 Apr 12;12(2):107-120. doi: 10.3920/BM2020.0132. Epub 2021 Apr 1. Benef Microbes. 2021. PMID: 33789552 Review. - Bacteriophages of the Human Gut: The "Known Unknown" of the Microbiome.
Shkoporov AN, Hill C. Shkoporov AN, et al. Cell Host Microbe. 2019 Feb 13;25(2):195-209. doi: 10.1016/j.chom.2019.01.017. Cell Host Microbe. 2019. PMID: 30763534 Review. - Phages and Human Health: More Than Idle Hitchhikers.
Lawrence D, Baldridge MT, Handley SA. Lawrence D, et al. Viruses. 2019 Jun 27;11(7):587. doi: 10.3390/v11070587. Viruses. 2019. PMID: 31252683 Free PMC article. Review. - Paving the Way to Unveil the Diversity and Evolution of Phage Genomes.
Reyes A, Vives MJ. Reyes A, et al. Viruses. 2020 Aug 19;12(9):905. doi: 10.3390/v12090905. Viruses. 2020. PMID: 32824934 Free PMC article.
Cited by
- High throughput sequencing provides exact genomic locations of inducible prophages and accurate phage-to-host ratios in gut microbial strains.
Zünd M, Ruscheweyh HJ, Field CM, Meyer N, Cuenca M, Hoces D, Hardt WD, Sunagawa S. Zünd M, et al. Microbiome. 2021 Mar 29;9(1):77. doi: 10.1186/s40168-021-01033-w. Microbiome. 2021. PMID: 33781335 Free PMC article. - The human virome: assembly, composition and host interactions.
Liang G, Bushman FD. Liang G, et al. Nat Rev Microbiol. 2021 Aug;19(8):514-527. doi: 10.1038/s41579-021-00536-5. Epub 2021 Mar 30. Nat Rev Microbiol. 2021. PMID: 33785903 Free PMC article. Review. - VirSorter: mining viral signal from microbial genomic data.
Roux S, Enault F, Hurwitz BL, Sullivan MB. Roux S, et al. PeerJ. 2015 May 28;3:e985. doi: 10.7717/peerj.985. eCollection 2015. PeerJ. 2015. PMID: 26038737 Free PMC article. - Efficacy of phage cocktail AB-SA01 therapy in diabetic mouse wound infections caused by multidrug-resistant Staphylococcus aureus.
Kifelew LG, Warner MS, Morales S, Vaughan L, Woodman R, Fitridge R, Mitchell JG, Speck P. Kifelew LG, et al. BMC Microbiol. 2020 Jul 9;20(1):204. doi: 10.1186/s12866-020-01891-8. BMC Microbiol. 2020. PMID: 32646376 Free PMC article. - Standard Bacteriophage Purification Procedures Cause Loss in Numbers and Activity.
Carroll-Portillo A, Coffman CN, Varga MG, Alcock J, Singh SB, Lin HC. Carroll-Portillo A, et al. Viruses. 2021 Feb 20;13(2):328. doi: 10.3390/v13020328. Viruses. 2021. PMID: 33672780 Free PMC article.
References
- Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. General nature of the genetic code for proteins. Nature. 1961;192:1227–1232. - PubMed
- Carins J, Stent GS, Watson JD. Phage and the Origins of Molecular Biology. Cold Spring Harbor Laboratory Press; Plainview, NY: 1992.
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM007200/GM/NIGMS NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- R01 DK030292/DK/NIDDK NIH HHS/United States
- R01 DK070977/DK/NIDDK NIH HHS/United States
- DK70977/DK/NIDDK NIH HHS/United States
- GM095384/GM/NIGMS NIH HHS/United States
- R01 GM095384/GM/NIGMS NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
- T32 GM007200/GM/NIGMS NIH HHS/United States
- DK30292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous