Virtual nanoscopy: generation of ultra-large high resolution electron microscopy maps - PubMed (original) (raw)
Virtual nanoscopy: generation of ultra-large high resolution electron microscopy maps
Frank G A Faas et al. J Cell Biol. 2012.
Abstract
A key obstacle in uncovering the orchestration between molecular and cellular events is the vastly different length scales on which they occur. We describe here a methodology for ultrastructurally mapping regions of cells and tissue as large as 1 mm(2) at nanometer resolution. Our approach employs standard transmission electron microscopy, rapid automated data collection, and stitching to create large virtual slides. It greatly facilitates correlative light-electron microscopy studies to relate structure and function and provides a genuine representation of ultrastructural events. The method is scalable as illustrated by slides up to 281 gigapixels in size. Here, we applied virtual nanoscopy in a correlative light-electron microscopy study to address the role of the endothelial glycocalyx in protein leakage over the glomerular filtration barrier, in an immunogold labeling study of internalization of oncolytic reovirus in human dendritic cells, in a cryo-electron microscopy study of intact vitrified mouse embryonic cells, and in an ultrastructural mapping of a complete zebrafish embryo slice.
Figures
Figure 1.
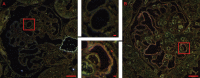
Reflection contrast microscopy (RCM) images of DAB peroxidase–stained mice glomeruli. Images of a control mouse (A) and a hyaluronidase-treated mouse (B) are shown. The glomerular capillaries of the hyaluronidase-treated mice are characterized by a bright pink color.
Figure 2.
Overlay of RCM and TEM images of two sequential 60-nm sections of a mouse glomerulus. Albumin was localized by DAB peroxidase staining, and is colored yellow in the RCM image, whereas the DAB precipitate is black in the TEM image. Bars: (A) 10 µm; (B) 1 µm, (C) 100 nm. The online slide can be found in the JCB DataViewer.
Figure 3.
Immunogold-labeled section of reovirus-infected human dendritic cells. The 10-nm gold particles were automatically detected and highlighted by green Gaussian profiles with 235 nm FWHM so that their locations show up at all magnifications. A section through ∼84 cells is displayed in A. Bar, 2 µm. (B) High magnification snapshot of an infected cell with viruses at its surface. (C) Snapshot of a cell that internalized the viruses. Bars (B and C) 100 nm. The online slide can be found in the JCB DataViewer.
Figure 4.
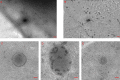
Cryo-electron microscopy virtual slide of a thin part of an unstained vitrified mouse embryonic fibroblast. (A) Low magnification overview of an area of 19 × 13 µm2. (B) The same region background corrected for the varying thickness of the cell. (C–E) High magnification snapshots of a microsome (C), a mitochondrion (D), and a multilamellar body as well as microtubules (E). Bars (A and B) 1 µm; (C–E) 100 nm. The online slide can be found in the JCB DataViewer.
Figure 5.
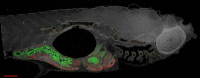
Overview and high magnification snapshots of a 281-Gpixel image of a 5-dpf zebrafish embryo sagittal section. The overview is shown in A with a legend that shows the colors used to mark 11 different tissues. High magnification snapshots are shown in B–E. (B) microvilli in the intestine, (C) cross sections of cilia of the olfactory pit, (D) notochord caveolae, and (E) junctional complex with two desmosomes. Bars (A) 100 µm; (B–E) 100 nm. The online slide can be found in the JCB DataViewer.
Figure 6.
Distribution of bare vs. membrane-wrapped mitochondria within selected tissues of the zebrafish virtual slide superimposed on a contrast-inverted overview. The distribution of the wrapped mitochondria is shown in red, and the bare mitochondria are shown in green. The locations of the mitochondria within the liver, yolk, pancreas, cartilage, notochord, and intestine are shown. Bar, 100 µm.
Figure 7.
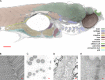
ER wrapping of mitochondria within the liver of the zebrafish embryo. The main panel shows a subregion of Fig. 5, in which the liver is highlighted. A–H show examples of mitochondria (light blue) wrapped with rER (yellow) or cisternae that lacked ribosomes (orange, E and F). Lysosomes (green) are often found nearby the enwrapped mitochondrion (A–D). The location of the mitochondria of A–H within the liver is marked with red letters in the main panel. Bar: (main panel) 10 µm; (A) 1 µm (bar in A represents magnification for B–H).
Figure 8.
Phagocytosis within the yolk region of the zebrafish embryo. The main panel highlights the left part of the yolk region of the sagittal section shown in Fig. 5. The positions of the close-ups shown in A–E and H–J are marked with red letters in the main panel; positions F and G are found in a neighboring part of the yolk. G–J show mitochondria in close contact to an rER cisterna. C–E show different stages of lysosomal wrapping; A, B, F, and G show autophagosomes. Bar: (main panel) 10 µm; (A) 1 µm (bar in A represents magnification for B–J).
Comment in
- The JCB DataViewer scales up.
Williams EH, Carpentier P, Misteli T. Williams EH, et al. J Cell Biol. 2012 Aug 6;198(3):271-2. doi: 10.1083/jcb.201207117. J Cell Biol. 2012. PMID: 22869591 Free PMC article.
Similar articles
- Detection and Characterization of Extracellular Vesicles by Transmission and Cryo-Transmission Electron Microscopy.
Cizmar P, Yuana Y. Cizmar P, et al. Methods Mol Biol. 2017;1660:221-232. doi: 10.1007/978-1-4939-7253-1_18. Methods Mol Biol. 2017. PMID: 28828660 - Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells.
van Driel LF, Valentijn JA, Valentijn KM, Koning RI, Koster AJ. van Driel LF, et al. Eur J Cell Biol. 2009 Nov;88(11):669-84. doi: 10.1016/j.ejcb.2009.07.002. Epub 2009 Sep 2. Eur J Cell Biol. 2009. PMID: 19726102 - Differentiation and Characterization of Excitatory and Inhibitory Synapses by Cryo-electron Tomography and Correlative Microscopy.
Tao CL, Liu YT, Sun R, Zhang B, Qi L, Shivakoti S, Tian CL, Zhang P, Lau PM, Zhou ZH, Bi GQ. Tao CL, et al. J Neurosci. 2018 Feb 7;38(6):1493-1510. doi: 10.1523/JNEUROSCI.1548-17.2017. Epub 2018 Jan 8. J Neurosci. 2018. PMID: 29311144 Free PMC article. - Electron tomography in plant cell biology.
Otegui MS, Pennington JG. Otegui MS, et al. Microscopy (Oxf). 2019 Feb 1;68(1):69-79. doi: 10.1093/jmicro/dfy133. Microscopy (Oxf). 2019. PMID: 30452668 Review. - Contribution of high-resolution correlative imaging techniques in the study of the liver sieve in three-dimensions.
Braet F, Wisse E, Bomans P, Frederik P, Geerts W, Koster A, Soon L, Ringer S. Braet F, et al. Microsc Res Tech. 2007 Mar;70(3):230-42. doi: 10.1002/jemt.20408. Microsc Res Tech. 2007. PMID: 17279510 Review.
Cited by
- Description and functional validation of human enteroendocrine cell sensors.
Beumer J, Geurts MH, Geurts V, Andersson-Rolf A, Akkerman N, Völlmy F, Krueger D, Busslinger GA, Martínez-Silgado A, Boot C, Yousef Yengej FA, Puschhof J, Van de Wetering WJ, Knoops K, López-Iglesias C, Peters PJ, Vivié JA, Mooijman D, van Es JH, Clevers H. Beumer J, et al. Science. 2024 Oct 18;386(6719):341-348. doi: 10.1126/science.adl1460. Epub 2024 Oct 17. Science. 2024. PMID: 39418382 Free PMC article. - Tuft cells act as regenerative stem cells in the human intestine.
Huang L, Bernink JH, Giladi A, Krueger D, van Son GJF, Geurts MH, Busslinger G, Lin L, Begthel H, Zandvliet M, Buskens CJ, Bemelman WA, López-Iglesias C, Peters PJ, Clevers H. Huang L, et al. Nature. 2024 Oct;634(8035):929-935. doi: 10.1038/s41586-024-07952-6. Epub 2024 Oct 2. Nature. 2024. PMID: 39358509 Free PMC article. - Systems mapping of bidirectional endosomal transport through the crowded cell.
Jongsma MLM, Bakker N, Voortman LM, Koning RI, Bos E, Akkermans JJLL, Janssen L, Neefjes J. Jongsma MLM, et al. Curr Biol. 2024 Oct 7;34(19):4476-4494.e11. doi: 10.1016/j.cub.2024.08.026. Epub 2024 Sep 13. Curr Biol. 2024. PMID: 39276769 Free PMC article. - The ORP9-ORP11 dimer promotes sphingomyelin synthesis.
Cabukusta B, Borst Pauwels S, Akkermans JJLL, Blomberg N, Mulder AA, Koning RI, Giera M, Neefjes J. Cabukusta B, et al. Elife. 2024 Aug 6;12:RP91345. doi: 10.7554/eLife.91345. Elife. 2024. PMID: 39106189 Free PMC article. - Bacteriophage ISP eliminates Staphylococcus aureus in planktonic phase, but not in the various stages of the biofilm cycle.
Verheul M, Mulder AA, van Dun SCJ, Merabishvili M, Nelissen RGHH, de Boer MGJ, Pijls BG, Nibbering PH. Verheul M, et al. Sci Rep. 2024 Jun 22;14(1):14374. doi: 10.1038/s41598-024-65143-9. Sci Rep. 2024. PMID: 38909125 Free PMC article.
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources