Biogenesis of telomerase ribonucleoproteins - PubMed (original) (raw)
Review
Biogenesis of telomerase ribonucleoproteins
Emily D Egan et al. RNA. 2012 Oct.
Abstract
Telomerase adds simple-sequence repeats to the ends of linear chromosomes to counteract the loss of end sequence inherent in conventional DNA replication. Catalytic activity for repeat synthesis results from the cooperation of the telomerase reverse transcriptase protein (TERT) and the template-containing telomerase RNA (TER). TERs vary widely in sequence and structure but share a set of motifs required for TERT binding and catalytic activity. Species-specific TER motifs play essential roles in RNP biogenesis, stability, trafficking, and regulation. Remarkably, the biogenesis pathways that generate mature TER differ across eukaryotes. Furthermore, the cellular processes that direct the assembly of a biologically functional telomerase holoenzyme and its engagement with telomeres are evolutionarily varied and regulated. This review highlights the diversity of strategies for telomerase RNP biogenesis, RNP assembly, and telomere recruitment among ciliates, yeasts, and vertebrates and suggests common themes in these pathways and their regulation.
Figures
FIGURE 1.
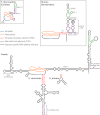
Diagram of TER secondary structures highlighting functional motifs. The template, pseudoknot, TBE, and STE are common to ciliate, yeast, and vertebrate TERs. The STE is distal stem–loop IV in T. thermophila, conserved region 4/5 (CR4/5) in human, and a three-way helix junction in yeasts. Species-specific RNP stability elements recruit p65 in T. thermophila, H/ACA proteins in human, and Sm proteins in yeasts (also Ku in Saccharomyces). The binding sites for holoenzyme proteins that do not affect RNA stability, namely Est1 in yeasts and the CAB box-binding protein WDR79/TCAB1 in humans, are indicated. T. thermophila TER also contains a template recognition element (TRE) that contributes to template utilization.
FIGURE 2.
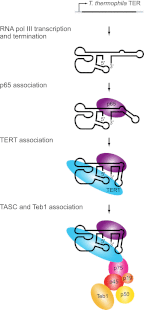
T. thermophila telomerase RNP biogenesis. T. thermophila TER is transcribed by RNA polymerase III. The binding of p65 stabilizes a kink in stem IV that optimally positions loop IV for TERT binding. The p65-TER–TERT ternary complex then interacts with a complex of p75, p50, p45, and p19 that recruits the single-stranded telomeric DNA-binding protein Teb1.
FIGURE 3.
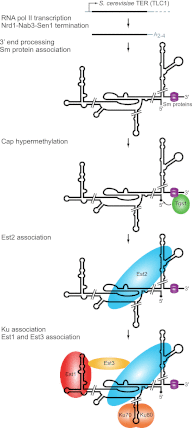
S. cerevisiae telomerase RNP biogenesis. S. cerevisiae TLC1 is transcribed by RNA polymerase II, and transcription is terminated by the Nrd1–Nab3–Sen1 pathway. The 3′ end is processed by the nuclear exosome and TRAMP complex, leaving a short adenosine-rich tail that is ultimately removed. Sm protein binding near the 3′ end stabilizes the RNA and recruits the cap hypermethylase Tgs1, which modifies the 5′ cap to TMG, indicated by a diamond. The TERT subunit Est2 binds directly to TLC1. The Ku heterodimer recognizes a distinct binding site on TLC1 to promote RNP accumulation and nuclear import. The regulatory subunit Est1 can bind directly to TLC1 and with Est3 stimulates telomerase function at telomeres.
FIGURE 4.
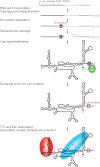
S. pombe telomerase RNP biogenesis. S. pombe TER1 is transcribed by RNA polymerase II as a precursor that includes a downstream intron and exon. TER1 secondary structure has yet to be experimentally validated; it is illustrated here as similar to other yeast TERs. Sm proteins assemble on the precursor near the mature TER1 3′ end. Spliceosomal cleavage at the 5′ splice site releases mature TER1, which escapes ligation to the downstream exon. Sm proteins then recruit the cap hypermethylase Tgs1, which modifies the 5′ cap to TMG, indicated by a diamond. Sm proteins are then replaced by Lsm proteins that protect the 3′ end from degradation, followed by assembly of Est1 and the TERT subunit Trt1.
FIGURE 5.
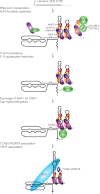
Human telomerase RNP biogenesis. The human TER, hTR, is transcribed by RNA polymerase II as a precursor that cotranscriptionally assembles with the H/ACA protein heterotrimer of dyskerin, NHP2, and NOP10 bound to the H/ACA RNP assembly chaperone NAF1. This process is aided by the dyskerin chaperone SHQ1 and a complex of NUFIP and the helicases RUVBL1 and RUVBL2. NUFIP interacts with NHP2, and the RUVBL1/RUVBL2 heterodimer interacts with dyskerin to promote H/ACA RNP assembly. The hTR precursor transcript is then processed at its 3′ end by an unknown mechanism. The G-quadruplex structure that can form near the 5′ end, represented by a wavy line, is resolved by the helicase DHX36. Then, NAF1 is exchanged for the mature H/ACA RNP protein GAR1, and sTGS1 modifies the 5′ cap to TMG, indicated by a diamond. TCAB1 and TERT bind to hTR in the active telomerase holoenzyme.
Similar articles
- Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability.
Robart AR, O'Connor CM, Collins K. Robart AR, et al. RNA. 2010 Mar;16(3):563-71. doi: 10.1261/rna.1936410. Epub 2010 Jan 27. RNA. 2010. PMID: 20106956 Free PMC article. - The architecture of Tetrahymena telomerase holoenzyme.
Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. Jiang J, et al. Nature. 2013 Apr 11;496(7444):187-92. doi: 10.1038/nature12062. Epub 2013 Apr 3. Nature. 2013. PMID: 23552895 Free PMC article. - Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65.
Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. Singh M, et al. Mol Cell. 2012 Jul 13;47(1):16-26. doi: 10.1016/j.molcel.2012.05.018. Epub 2012 Jun 14. Mol Cell. 2012. PMID: 22705372 Free PMC article. - Evolutionary perspectives of telomerase RNA structure and function.
Podlevsky JD, Chen JJ. Podlevsky JD, et al. RNA Biol. 2016 Aug 2;13(8):720-32. doi: 10.1080/15476286.2016.1205768. Epub 2016 Jun 30. RNA Biol. 2016. PMID: 27359343 Free PMC article. Review. - Telomerase Mechanism of Telomere Synthesis.
Wu RA, Upton HE, Vogan JM, Collins K. Wu RA, et al. Annu Rev Biochem. 2017 Jun 20;86:439-460. doi: 10.1146/annurev-biochem-061516-045019. Epub 2017 Jan 30. Annu Rev Biochem. 2017. PMID: 28141967 Free PMC article. Review.
Cited by
- SMG6 regulates DNA damage and cell survival in Hippo pathway kinase LATS2-inactivated malignant mesothelioma.
Suzuki K, Tange M, Yamagishi R, Hanada H, Mukai S, Sato T, Tanaka T, Akashi T, Kadomatsu K, Maeda T, Miida T, Takeuchi I, Murakami H, Sekido Y, Murakami-Tonami Y. Suzuki K, et al. Cell Death Discov. 2022 Nov 5;8(1):446. doi: 10.1038/s41420-022-01232-w. Cell Death Discov. 2022. PMID: 36335095 Free PMC article. - Synonymous Mutation in DKC1 Causes Telomerase RNA Insufficiency Manifesting as Familial Pulmonary Fibrosis.
Gaysinskaya V, Stanley SE, Adam S, Armanios M. Gaysinskaya V, et al. Chest. 2020 Dec;158(6):2449-2457. doi: 10.1016/j.chest.2020.07.025. Epub 2020 Jul 22. Chest. 2020. PMID: 32710892 Free PMC article. - Diverse mechanisms for spliceosome-mediated 3' end processing of telomerase RNA.
Kannan R, Helston RM, Dannebaum RO, Baumann P. Kannan R, et al. Nat Commun. 2015 Jan 19;6:6104. doi: 10.1038/ncomms7104. Nat Commun. 2015. PMID: 25598145 Free PMC article. - Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies.
Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, Hockemeyer D, Collins K. Vogan JM, et al. Elife. 2016 Aug 15;5:e18221. doi: 10.7554/eLife.18221. Elife. 2016. PMID: 27525486 Free PMC article. - High-throughput telomere length measurement at nucleotide resolution using the PacBio high fidelity sequencing platform.
Tham CY, Poon L, Yan T, Koh JYP, Ramlee MK, Teoh VSI, Zhang S, Cai Y, Hong Z, Lee GS, Liu J, Song HW, Hwang WYK, Teh BT, Tan P, Xu L, Koh AS, Osato M, Li S. Tham CY, et al. Nat Commun. 2023 Jan 17;14(1):281. doi: 10.1038/s41467-023-35823-7. Nat Commun. 2023. PMID: 36650155 Free PMC article.
References
- Autexier C, Greider CW 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev 9: 2227–2239 - PubMed
- Autexier C, Lue NF 2006. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem 75: 493–517 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM054198/GM/NIGMS NIH HHS/United States
- R56 HL079585/HL/NHLBI NIH HHS/United States
- R01 HL079585/HL/NHLBI NIH HHS/United States
- HL079585/HL/NHLBI NIH HHS/United States
- CA009041/CA/NCI NIH HHS/United States
- T32 CA009041/CA/NCI NIH HHS/United States
- R01 GM054198/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources