Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis - PubMed (original) (raw)
doi: 10.1371/journal.pone.0043090. Epub 2012 Aug 15.
Valeria Visconte, Anna M Jankowska, Hideki Makishima, Christine L O'Keefe, Paul Elson, Yingchun Han, Fred H Hsieh, Mikkael A Sekeres, Raghuveer Singh Mali, Matt Kalaycio, Alan E Lichtin, Anjali S Advani, Hien K Duong, Edward Copelan, Reuben Kapur, Sara T Olalla Saad, Jaroslaw P Maciejewski, Ramon V Tiu
Affiliations
- PMID: 22905207
- PMCID: PMC3419680
- DOI: 10.1371/journal.pone.0043090
Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis
Fabiola Traina et al. PLoS One. 2012.
Abstract
We hypothesized that analysis of single nucleotide polymorphism arrays (SNP-A) and new molecular defects may provide new insight in the pathogenesis of systemic mastocytosis (SM). SNP-A karyotyping was applied to identify recurrent areas of loss of heterozygosity and bidirectional sequencing was performed to evaluate the mutational status of TET2, DNMT3A, ASXL1, EZH2, IDH1/IDH2 and the CBL gene family. Overall survival (OS) was analyzed using the Kaplan-Meier method. We studied a total of 26 patients with SM. In 67% of SM patients, SNP-A karyotyping showed new chromosomal abnormalities including uniparental disomy of 4q and 2p spanning TET2/KIT and DNMT3A. Mutations in TET2, DNMT3A, ASXL1 and CBL were found in 23%, 12%, 12%, and 4% of SM patients, respectively. No mutations were observed in EZH2 and IDH1/IDH2. Significant differences in OS were observed for SM mutated patients grouped based on the presence of combined TET2/DNMT3A/ASXL1 mutations independent of KIT (P = 0.04) and sole TET2 mutations (P<0.001). In conclusion, TET2, DNMT3A and ASXL1 mutations are also present in mastocytosis and these mutations may affect prognosis, as demonstrated by worse OS in mutated patients.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
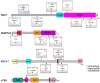
Figure 1. Single nucleotide polymorphism array-based karyotyping (SNP-A) of mastocytosis patients.
(A) Overview of all genetic aberrations found by SNP-A analysis in patients with systemic mastocytosis. Green represents gain, red represents loss, black represents somatic uniparental disomy (UPD). UPD involving the KIT and TET2 genes on chromosome 4q and UPD involving the DNMT3A gene on chromosome 2p were noted in one patient each, as indicated. (B) Representative SNP-A analysis of loss of heterozygosity (LOH), UPD, and gain by Genotyping Console v3.0. The top track of each panel shows LOH. The second track shows raw copy number for each SNP along the chromosome, while the third track shows allele calls (AA, AB, BB). Each region of genomic change is indicated by vertical black bars.
Figure 2. Localization of mutations identified in systemic mastocytosis.
In a cohort of 26 patients with systemic mastocytosis, 14 mutations were identified. Genomic sequencing of protein-coding regions and splice sites revealed missense (black), nonsense (orange), and frameshift mutations (blue) in TET2, DNMT3A, ASXL1, and CBL. Most mutations were found in conserved domains and specific known conserved motifs and domains are shown for each protein: cysteine-rich region (C-rich-), double strand â helix (DSBH), PWWP domain (characterized by the presence of a highly conserved proline–tryptophan–tryptophan–proline motif), ADD (ATRX, DNMT3, and DNMT3L)-type zinc finger (ZNF) domain, methyltransferase (MTase) domain, amino-terminal ASX homology (ASXN) region, ASXM domain, nuclear receptor coregulator binding (NR box) motifs, carboxyterminal plant homeodomain (PHD) domain, tyrosine kinase binding (TKB) domain, linker sequence (L), RF domain (RF), proline-rich region (PPP), and leucine zipper LZ/ubiquitin-associated domain (UBA). Two changes occurred in a homozygous state, as indicated by the symbol # and the others in heterozygous state.
Figure 3. Kaplan-Meier survival curves estimated according to presence of specific mutations or accumulation of several mutations in patients with systemic mastocytosis.
Differences in OS for SM patients are shown (A-B). For each group number of analyzed cases and P value are presented, respectively.
Similar articles
- ASXL1 but not TET2 mutations adversely impact overall survival of patients suffering systemic mastocytosis with associated clonal hematologic non-mast-cell diseases.
Damaj G, Joris M, Chandesris O, Hanssens K, Soucie E, Canioni D, Kolb B, Durieu I, Gyan E, Livideanu C, Chèze S, Diouf M, Garidi R, Georgin-Lavialle S, Asnafi V, Lhermitte L, Lavigne C, Launay D, Arock M, Lortholary O, Dubreuil P, Hermine O. Damaj G, et al. PLoS One. 2014 Jan 21;9(1):e85362. doi: 10.1371/journal.pone.0085362. eCollection 2014. PLoS One. 2014. PMID: 24465546 Free PMC article. - TET2, ASXL1, IDH1, IDH2, and c-CBL genes in JAK2- and MPL-negative myeloproliferative neoplasms.
Martínez-Avilés L, Besses C, Álvarez-Larrán A, Torres E, Serrano S, Bellosillo B. Martínez-Avilés L, et al. Ann Hematol. 2012 Apr;91(4):533-41. doi: 10.1007/s00277-011-1330-0. Epub 2011 Sep 9. Ann Hematol. 2012. PMID: 21904853 - Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms.
Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, Cervera N, Gelsi-Boyer V, Arnoulet C, Gisserot O, Verrot D, Slama B, Vey N, Mozziconacci MJ, Birnbaum D, Murati A. Brecqueville M, et al. Genes Chromosomes Cancer. 2012 Aug;51(8):743-55. doi: 10.1002/gcc.21960. Epub 2012 Apr 9. Genes Chromosomes Cancer. 2012. PMID: 22489043 - Epigenetic Changes in Neoplastic Mast Cells and Potential Impact in Mastocytosis.
Reszka E, Jabłońska E, Wieczorek E, Valent P, Arock M, Nilsson G, Nedoszytko B, Niedoszytko M. Reszka E, et al. Int J Mol Sci. 2021 Mar 15;22(6):2964. doi: 10.3390/ijms22062964. Int J Mol Sci. 2021. PMID: 33803981 Free PMC article. Review. - Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1.
Tefferi A. Tefferi A. Leukemia. 2010 Jun;24(6):1128-38. doi: 10.1038/leu.2010.69. Epub 2010 Apr 29. Leukemia. 2010. PMID: 20428194 Free PMC article. Review.
Cited by
- Detection of KIT Mutations in Systemic Mastocytosis: How, When, and Why.
Cilloni D, Maffeo B, Savi A, Danzero AC, Bonuomo V, Fava C. Cilloni D, et al. Int J Mol Sci. 2024 Oct 10;25(20):10885. doi: 10.3390/ijms252010885. Int J Mol Sci. 2024. PMID: 39456668 Free PMC article. Review. - Decoding Clonal Hematopoiesis: Emerging Themes and Novel Mechanistic Insights.
Pendse S, Loeffler D. Pendse S, et al. Cancers (Basel). 2024 Jul 24;16(15):2634. doi: 10.3390/cancers16152634. Cancers (Basel). 2024. PMID: 39123361 Free PMC article. Review. - Avapritinib treatment of aggressive systemic mastocytosis with a novel KIT exon 17 mutation.
Sandow L, Town A, Heinrich MC. Sandow L, et al. Leuk Res Rep. 2023 Dec 30;21:100409. doi: 10.1016/j.lrr.2023.100409. eCollection 2024. Leuk Res Rep. 2023. PMID: 38273969 Free PMC article. - A multicenter retrospective comparison between systemic mastocytosis with t(8;21) AML and KIT mutant t(8;21) AML.
Zhang Z, Yin J, Lian G, Bao X, Hu M, Liu Z, Yu Y, Mi R, Zuo Y, Shi P, Zheng W, Jiang Q, Chao H, Xiao P, Yu W, Han Y, Wu Y, Zeng Y, Wu D, Yang X, Chen S. Zhang Z, et al. Blood Adv. 2024 Feb 27;8(4):889-894. doi: 10.1182/bloodadvances.2023012006. Blood Adv. 2024. PMID: 38170739 Free PMC article. No abstract available. - CDK4/CDK6 Inhibitors Synergize with Midostaurin, Avapritinib, and Nintedanib in Inducing Growth Inhibition in KIT D816V+ Neoplastic Mast Cells.
Schneeweiss-Gleixner M, Filik Y, Stefanzl G, Berger D, Sadovnik I, Bauer K, Smiljkovic D, Eisenwort G, Witzeneder N, Greiner G, Hoermann G, Schiefer AI, Schwaab J, Jawhar M, Reiter A, Sperr WR, Arock M, Valent P, Gleixner KV. Schneeweiss-Gleixner M, et al. Cancers (Basel). 2022 Jun 23;14(13):3070. doi: 10.3390/cancers14133070. Cancers (Basel). 2022. PMID: 35804842 Free PMC article.
References
- Valent P, Horny HP, Escribano L, Longley BJ, Li CY, et al. (2001) Diagnostic criteria and classification of mastocytosis: a consensus proposal. Leuk Res 25: 603–625. - PubMed
- Valent P, Akin C, Escribano L, Fodinger M, Hartmann K, et al. (2007) Standards and standardization in mastocytosis: consensus statements on diagnostics, treatment recommendations and response criteria. Eur J Clin Invest 37: 435–453. - PubMed
- Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, et al. (1999) Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13). Blood 94: 2333–2342. - PubMed
- Tefferi A, Pardanani A (2004) Clinical, genetic, and therapeutic insights into systemic mast cell disease. Curr Opin Hematol 11: 58–64. - PubMed
- Horny HP, Metcalfe DD, Bennett BD, Bain BJ, Akin C, et al. (2008) Mastocytosis. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, et al.., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 54–63.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous