Membrane binding and self-association of the epsin N-terminal homology domain - PubMed (original) (raw)
Membrane binding and self-association of the epsin N-terminal homology domain
Chun-Liang Lai et al. J Mol Biol. 2012.
Abstract
Epsin possesses a conserved epsin N-terminal homology (ENTH) domain that acts as a phosphatidylinositol 4,5-bisphosphate-lipid-targeting and membrane-curvature-generating element. Upon binding phosphatidylinositol 4,5-bisphosphate, the N-terminal helix (H(0)) of the ENTH domain becomes structured and aids in the aggregation of ENTH domains, which results in extensive membrane remodeling. In this article, atomistic and coarse-grained (CG) molecular dynamics (MD) simulations are used to investigate the structure and the stability of ENTH domain aggregates on lipid bilayers. EPR experiments are also reported for systems composed of different ENTH-bound membrane morphologies, including membrane vesicles as well as preformed membrane tubules. The EPR data are used to help develop a molecular model of ENTH domain aggregates on preformed lipid tubules that are then studied by CG MD simulation. The combined computational and experimental approach suggests that ENTH domains exist predominantly as monomers on vesiculated structures, while ENTH domains self-associate into dimeric structures and even higher-order oligomers on the membrane tubes. The results emphasize that the arrangement of ENTH domain aggregates depends strongly on whether the local membrane curvature is isotropic or anisotropic. The molecular mechanism of ENTH-domain-induced membrane vesiculation and tubulation and the implications of the epsin's role in clathrin-mediated endocytosis resulting from the interplay between ENTH domain membrane binding and ENTH domain self-association are also discussed.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
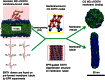
Graphical abstract
Fig. 1
(a) Atomistic MD simulation of the ENTH domain in a lipid bilayer. Representative snapshots are taken from MD simulation of the complex composed of an ENTH domain and a bilayer containing one PIP2 molecule. The ENTH domain is represented as a red ribbon. Lipid headgroups are shown in blue and lipid tails are shown in green. Two black arrows indicate the locations of the R114 loop and H0 helix site. The PIP2-bound site of the ENTH domain is indicated by a cyan arrow. Red points represent oxygen atoms of waters in the MD system. (b) The average insertion depth of Cα from residues 4 to 14 of helix H0 is shown. Depth is measured parallel to the membrane normal with respect to the lipid bilayer phosphate groups. (c) Distribution of the center of mass of H0 with respect to the lipid bilayer phosphate groups as was calculated from the last 50‐ns trajectory of MD simulation. (d) Close-up view of the ENTH domain membrane-interacting motifs and the binding site of the PIP2 headgroup to the ENTH domain. A gray transparent shadow is used to illustrate the location of lipid bilayer. The protein backbone is shown as a red ribbon, except for the R114 loop and H0 site, which are highlighted in blue. Residue R114 and a PIP2 molecule are explicitly included, shown in sphere representation. The eight residues indicated in Ford et al. that directly interact with the PIP2 headgroup are shown in a blue ball-and-stick representation. (e) Accessibility measurements indicate the formation of an amphipathic helical structure in the N-terminus of the epsin 1 ENTH domain. (e) shows the contrast parameter Φ as a function of residue number. Note that higher values of Φ correspond to deeper Cα insertion in (b). The period of oscillation is indicative of helical structure as indicated by the sinusoidal line drawn with an ideal periodicity of 3.6 amino acids. (f) Local maxima (magenta circles) fall onto the hydrophobic face and local minima cluster on the hydrophilic side of the helical wheel representation shown in (f).
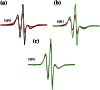
Fig. 2
EPR evidence for dimerization of the epsin 1 ENTH domain. The EPR spectra of 10R1 incubated with preformed tubules (red) or liposomes (black) are overlaid in (a). Preformed tubule‐bound 10R1 exhibits pronounced line broadening indicative of spin–spin interaction. To investigate the effect of spin interaction on the line shape, we incubated 10R1 with preformed tubules in the presence of twofold excess of unlabeled protein, resulting in a spectrum with reduced line broadening and increased amplitude [green trace in (b)]. (c) shows the analogous dilution experiment as shown in (b) with the exception that liposomes were used. The scan width for all spectra is 150 Gauss and all spectra are normalized to the same number of spins.
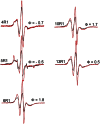
Fig. 3
EPR spectra for selected epsin 1 ENTH domain spin‐labeled derivatives bound to preformed tubules. The black spectra are obtained from proteins labeled at the indicated positions while the red spectra were obtained using the indicated spin‐labeled derivatives diluted with twofold excess of unlabeled protein.
Fig. 4
All-atom structure of the ENTH domain (a) and the 16‐site CG ENTH domain model (b). The two N-terminal amphipathic helix sites [CG-1 and CG-2 in (b)] and one R114 loop site [CG-10 in (b)] are shown in blue. The PIP2‐interacting site is shown in cyan. The CG site 5 [CG-5 in (b)] that consists of two solvent-exposed hydrophobic residues (V50 and V51) is shown in white. The rest of the domain is in red.
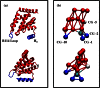
Fig. 5
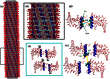
CG MD simulation system with an ENTH-lattice‐coated membrane tubule. (a) A membrane tubule is shown in gray. Red lines connect CG sites. Selected CG sites are shown in sphere representation. H0 and hydrophobic contact sites V50 and V51 are shown in blue and white spheres, respectively. (b) Close-up of (a). (c) Close-up ribbon view of the ENTH tetramer in (b). Four ENTH domains are shown in red ribbons, while H0 helices are shown in thicker blue ribbons. Hydrophobic contact patches, V50 and V51, are shown as white spheres. (d) Bottom view of (c), with only a dimer unit shown. L6 and M10 are shown in cyan spheres. The orange and light blue arrows represent the measured EPR distance (~ 13 Å) for L6 and M10, respectively. (e) Bottom view of (c). Offset packing between two dimers that brings the I13 and V14 region of H0 in close proximity as highlighted with a yellow arrow. I13 and V14 are shown in sphere representation. This is indicated by an EPR distance profile of 15 Å for I13 and 10 Å for V14. These distances are shorter than would be expected for an antiparallel dimer pairing.
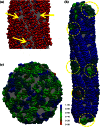
Fig. 6
(a) Close-up view of the ENTH domain lattice on a type A tube. Selected defect sites are shown with yellow arrows. Lipids are shown in gray. CG sites are shown as red spheres, except for H0 sites that are shown in blue spheres and PIP2 interaction sites that are shown in cyan. (b) Local orientational order of ENTH domain-bound membrane tube (type B tube). The simulation snapshot is colored to show the local orientational order in the neighborhood of each ENTH domain. The color scale reflects the average of the unsigned dot product between the H0 axes of neighboring ENTH domains within 5 nm. Blue corresponds to a high degree of local order, and red corresponds to local disorder. There is considerably more disorder for both the capping regions, and the defect sites are scattered across the tube in a nonuniform manner. Selected defect sites are highlighted in yellow dashed circles. (c) Local orientational order parameter color map of an ENTH domain-bound membrane vesicle. The representative simulation snapshot is taken from an equilibrated CG MD simulation.
Similar articles
- Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain.
Yoon Y, Tong J, Lee PJ, Albanese A, Bhardwaj N, Källberg M, Digman MA, Lu H, Gratton E, Shin YK, Cho W. Yoon Y, et al. J Biol Chem. 2010 Jan 1;285(1):531-40. doi: 10.1074/jbc.M109.068015. Epub 2009 Nov 1. J Biol Chem. 2010. PMID: 19880963 Free PMC article. - Complimentary action of structured and unstructured domains of epsin supports clathrin-mediated endocytosis at high tension.
Joseph JG, Osorio C, Yee V, Agrawal A, Liu AP. Joseph JG, et al. Commun Biol. 2020 Dec 8;3(1):743. doi: 10.1038/s42003-020-01471-6. Commun Biol. 2020. PMID: 33293652 Free PMC article. - ENTH/ANTH proteins and clathrin-mediated membrane budding.
Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. Legendre-Guillemin V, et al. J Cell Sci. 2004 Jan 1;117(Pt 1):9-18. doi: 10.1242/jcs.00928. J Cell Sci. 2004. PMID: 14657269 Review. - AP180 N-Terminal Homology (ANTH) and Epsin N-Terminal Homology (ENTH) Domains: Physiological Functions and Involvement in Disease.
Takatori S, Tomita T. Takatori S, et al. Adv Exp Med Biol. 2019;1111:55-76. doi: 10.1007/5584_2018_218. Adv Exp Med Biol. 2019. PMID: 29774507 Review.
Cited by
- Biophysical Screening Pipeline for Cryo-EM Grid Preparation of Membrane Proteins.
Niebling S, Veith K, Vollmer B, Lizarrondo J, Burastero O, Schiller J, Struve García A, Lewe P, Seuring C, Witt S, García-Alai M. Niebling S, et al. Front Mol Biosci. 2022 Jun 23;9:882288. doi: 10.3389/fmolb.2022.882288. eCollection 2022. Front Mol Biosci. 2022. PMID: 35813810 Free PMC article. - Coarse-Grained Models for Protein-Cell Membrane Interactions.
Bradley R, Radhakrishnan R. Bradley R, et al. Polymers (Basel). 2013;5(3):890-936. doi: 10.3390/polym5030890. Polymers (Basel). 2013. PMID: 26613047 Free PMC article. - Curvature-undulation coupling as a basis for curvature sensing and generation in bilayer membranes.
Bradley RP, Radhakrishnan R. Bradley RP, et al. Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):E5117-24. doi: 10.1073/pnas.1605259113. Epub 2016 Aug 16. Proc Natl Acad Sci U S A. 2016. PMID: 27531962 Free PMC article. - Phosphoinositides, kinases and adaptors coordinating endocytosis in Trypanosoma brucei.
Manna PT, Field MC. Manna PT, et al. Commun Integr Biol. 2016 Jan 19;8(6):e1082691. doi: 10.1080/19420889.2015.1082691. eCollection 2015 Nov-Dec. Commun Integr Biol. 2016. PMID: 27064836 Free PMC article. - Listeria monocytogenes Exploits Host Caveolin for Cell-to-Cell Spreading.
Dhanda AS, Yu C, Lulic KT, Vogl AW, Rausch V, Yang D, Nichols BJ, Kim SH, Polo S, Hansen CG, Guttman JA. Dhanda AS, et al. mBio. 2020 Jan 21;11(1):e02857-19. doi: 10.1128/mBio.02857-19. mBio. 2020. PMID: 31964732 Free PMC article.
References
- McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. - PubMed
- Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. - PubMed
- Itoh T., De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 2006;1761:897–912. - PubMed
- Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. - PubMed
- Itoh T., Takenawa T. Mechanisms of membrane deformation by lipid-binding domains. Prog. Lipid Res. 2009;48:298–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 092096/Wellcome Trust/United Kingdom
- R01 GM063796/GM/NIGMS NIH HHS/United States
- MC_U105178795/MRC_/Medical Research Council/United Kingdom
- R01 GM063915/GM/NIGMS NIH HHS/United States
- GM063915/GM/NIGMS NIH HHS/United States
- T32 GM067587/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources