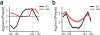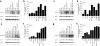Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange - PubMed (original) (raw)
Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange
Michaela Smolle et al. Nat Struct Mol Biol. 2012 Sep.
Abstract
Set2-mediated methylation of histone H3 Lys36 (H3K36) is a mark associated with the coding sequences of actively transcribed genes, but it has a negative role during transcription elongation. It prevents trans-histone exchange over coding regions and signals for histone deacetylation in the wake of RNA polymerase II (RNAPII) passage. We have found that in Saccharomyces cerevisiae the Isw1b chromatin-remodeling complex is specifically recruited to open reading frames (ORFs) by H3K36 methylation through the PWWP domain of its Ioc4 subunit in vivo and in vitro. Isw1b acts in conjunction with Chd1 to regulate chromatin structure by preventing trans-histone exchange from taking place over coding regions. In this way, Isw1b and Chd1 are important in maintaining chromatin integrity during transcription elongation by RNAPII.
Figures
Figure 1. Proteins associated with H3K36me3-containing mononucleosomes
(a) MudPIT mass spectrometry analysis of proteins co-immunoprecipitated with H3K36me3-containing mononucleosomes from wildtype yeast. IgG was used as a negative control. a Spectral count, total spectra matching peptides detected by tandem mass spectrometry for the indicated protein. b Sequence coverage, percentage of protein sequence represented in peptides identified by tandem mass spectrometry. (b) Subunit composition of Isw1 and Chd1 remodelers. Isw1 is the catalytic subunit of two chromatin-remodeling complexes: It associates with Ioc3 to form Isw1a, while Ioc2 and Ioc4 are part of the Isw1b complex. Chd1 is thought to exist largely as a monomer. (c) Schematic representations of the domain organizations for the Isw1 and Chd1 chromatin remodelers.
Figure 2. Ioc4 preferentially binds H3K36me3 nucleosomes
(a,b) Recombinant, reconstituted H3K36 MLA nucleosomes (5 fmol) were incubated with increasing concentrations (0, 3, 7, 10, 13, 17, 20 and 27 nM) of Ioc4 (a) or Ioc4 with its N-terminal PWWP domain deleted (Ioc4ΔPWWP) (b) and analyzed using EMSAs. Mononucleosome bands are indicated. (c) Quantitation of Ioc4 and Ioc4ΔPWWP binding to tri- and unmethylated H3K36 MLA mononucleosomes. Mononucleosome bands were quantitated for each lane. Lanes 2–8 were normalized against input lane 1, while lanes 10–16 were normalized to input lane 9. The percentage of nucleosomes bound by Ioc4 or Ioc4ΔPWWP was expressed as 100 – % mononucleosomes for each lane. Mean values ± SEM were plotted for at least three independent experiments. _P_-values were calculated using Student's t-test.
Figure 3. Deletion of SET2 abrogates localization of Ioc4 to coding regions
(a,b) ChIP-chip experiments were performed using yeast genomic tiling arrays. The log2 ratios of IP over input were subjected to average gene analysis (Supplementary Fig. 2a). Whole-genome average data were calculated and plotted as mean ± SEM. SE of mean are shown in grey and represent three independent experiments. The _T_ranscription _S_tart _S_ite (TSS) and _TE_rmination _S_ite (TES) are indicated. Flag-tagged Ioc4 (a) and Ioc3 (b) were immunoprecipitated from wildtype and set2Δ yeast strains.
Figure 4. ISW1 and CHD1 have overlapping functions during transcription within the Set2 pathway
(a,c,e,g) Total RNA for each strain was isolated and used for Northern blot analysis. The probes used were directed against the 3′ ends of the STE11 and PCA1 genes. ACT1 and rRNA were used as loading controls. The full-length (←) and cryptic transcripts (*) are indicated. (b,d,f,h) For each lane all cryptic transcript bands were quantitated and normalized against the ACT1 loading control. Total levels of cryptic transcription were expressed relative to set2Δ. Data were plotted as mean ± SEM for three independent experiments.
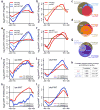
Figure 5. Deletion of ISW1 and CHD1 causes wide-spread intragenic transcription
(a) Heatmap of cryptic transcript genes in an isw1Δ chd1Δ mutant. Probe intensities for the 5′ and 3′ ends of all genes were determined. Resulting 3′/5′ ratios with a cutoff value of log2 > 0.5 were used to define sense cryptic transcripts (n=646). K-means clustering of gene expression profiles was used to identify antisense cryptic transcripts (n=962). Only cryptic transcript genes are shown. (b) Venn diagram showing the overlaps between genes displaying increased histone exchange over ORFs in set2Δ (mean log2 > 0; n=3,728) and genes with cryptic transcription in set2Δ (n=865) and/or isw1Δ chd1Δ (n=1,437). Cryptic transcript sets contain genes with sense and/or antisense cryptic transcripts, whereby 68 and 171 genes show intragenic initiation in both the sense and antisense direction for set2Δ and isw1Δ chd1Δ, respectively. For the purpose of direct comparison we selected cryptic transcript genes for set2Δ according to the same criteria applied to isw1Δ chd1Δ (a), rather than the previously published data.
Figure 6. Deletion of ISW1 and CHD1 increases histone exchange over 3′ ends of ORFs
(a–b,e–f) ChIP-chip experiments were performed using yeast genomic tiling arrays. Only genes known to display increased exchange over ORFs in a set2Δ mutant (n=3,728) (Supplementary Fig. 4a) were used for average gene analysis and plotted as mean ± SEM (grey) for three independent experiments. The _T_ranscription _S_tart _S_ite (TSS) and _TE_rmination _S_ite (TES) are indicated. (a–b) Histone exchange (Flag-H3/Myc-H3) was determined for wildtype, isw1Δ, chd1Δ, isw1Δ chd1Δ (a) and ioc4Δ (b) strains and presented as difference profiles for mutant (mut) over wildtype. (c–d) Genes were clustered into two groups based on their average histone exchange signals for mutant relative to wildtype profiles over ORFs (mean log2 < 0, mean log2 > 0). Venn diagrams show overlaps between genes displaying increased histone exchange over ORFs (mean log2 > 0) for the mutants indicated. (e–f) Average gene analysis for acetylated K56 (H3K56ac) immunoprecipitated from wildtype, isw1Δ, chd1Δ, isw1Δ chd1Δ (e) and the ISW1K227R catalytic mutant (f) strains. H3K56ac occupancy was normalized to histone H3 levels. Difference profiles are shown for mutant over wildtype. (g) Venn diagram showing the overlap between genes that exhibit cryptic transcription in an isw1Δ chd1Δ mutant (Fig. 5a,b) and genes that exhibit increased histone exchange over ORFs in an isw1Δ chd1Δ and set2Δ background. (h) Pearson correlation coefficients and _P_-values were calculated for histone exchange (a) and H3K56ac/H3 (e) profiles for each mutant background using R. (i–l) For whole-genome analysis genes (n=4,894) were clustered into two groups (< 10 mRNA per hour (4,250 genes), ≥ 10 mRNA per hour (644 genes)) based on transcription rates published by Holstege et al.. Average gene analysis for histone exchange (i–j) or H3K56ac/H3 (k–l) difference profiles clustered according to transcription rates are shown. Data were plotted for isw1Δ (i,k) or chd1Δ (j,l) relative to the wildtype.
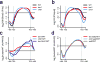
Figure 7. Deletion of ISW1 and CHD1 increases histone acetylation over coding regions
(a–d) ChIP-chip experiments were performed as described in Fig. 6. Only genes known to display increased exchange over ORFs in a set2Δ mutant (n=3,728) (Supplementary Fig. 4a) were used for average gene analysis and plotted as mean ± SEM (grey) for three independent experiments. The _T_ranscription _S_tart _S_ite (TSS) and _TE_rmination _S_ite (TES) are indicated. (a) Average gene analysis for H3K36me3 immunoprecipitated from wildtype, isw1Δ and chd1Δ yeast strains. (b) Average gene analysis for histone H3 immunoprecipitated from wildtype, isw1Δ and chd1Δ yeast strains. (c–d) Average gene analysis for acetylated H4 (AcH4) immunoprecipitated from wildtype, isw1Δ, chd1Δ, isw1Δ chd1Δ (c) as well as set2Δ, isw1Δ set2Δ and chd1Δ set2Δ (d) yeast strains. AcH4 occupancy was normalized to histone H3 levels. Difference profiles are shown for mutant over wildtype.
Figure 8. Isw1b and Chd1 maintain chromatin integrity over coding regions
Isw1b is recruited to ORFs by interaction of its Ioc4 subunit with H3K36me3, while Chd1 is thought to co-localize with RNAPII due to its interaction with known transcription elongation factors (grey). In the wildtype both remodelers cooperate to preserve chromatin structure by preventing histone exchange over ORFs. Deletion of ISW1, IOC4 or CHD1, or abrogation of Isw1 catalytic activity allows for trans-histone exchange to occur over coding sequences and results in increased levels of histone acetylation over ORFs. This perturbation of ORF chromatin structure exposes normally hidden cryptic promoters and results in the production of internal, cryptic transcripts.
Comment in
- Chromatin 'resetting' during transcription elongation: a central role for methylated H3K36.
Butler JS, Dent SY. Butler JS, et al. Nat Struct Mol Biol. 2012 Sep;19(9):863-4. doi: 10.1038/nsmb.2370. Nat Struct Mol Biol. 2012. PMID: 22955932 Free PMC article. - Chromatin: Lys36 sets limits for histone exchange.
Jones B. Jones B. Nat Rev Mol Cell Biol. 2012 Oct;13(10):602-3. doi: 10.1038/nrm3443. Epub 2012 Sep 20. Nat Rev Mol Cell Biol. 2012. PMID: 22992593 No abstract available.
Similar articles
- Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin.
Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Maltby VE, et al. Mol Cell Biol. 2012 Sep;32(17):3479-85. doi: 10.1128/MCB.00389-12. Epub 2012 Jul 2. Mol Cell Biol. 2012. PMID: 22751925 Free PMC article. - The yeast ISW1b ATP-dependent chromatin remodeler is critical for nucleosome spacing and dinucleosome resolution.
Eriksson PR, Clark DJ. Eriksson PR, et al. Sci Rep. 2021 Feb 18;11(1):4195. doi: 10.1038/s41598-021-82842-9. Sci Rep. 2021. PMID: 33602956 Free PMC article. - Widespread remodeling of mid-coding sequence nucleosomes by Isw1.
Tirosh I, Sigal N, Barkai N. Tirosh I, et al. Genome Biol. 2010;11(5):R49. doi: 10.1186/gb-2010-11-5-r49. Epub 2010 May 10. Genome Biol. 2010. PMID: 20459718 Free PMC article. - A site to remember: H3K36 methylation a mark for histone deacetylation.
Lee JS, Shilatifard A. Lee JS, et al. Mutat Res. 2007 May 1;618(1-2):130-4. doi: 10.1016/j.mrfmmm.2006.08.014. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17346757 Review. - ISWI complexes in Saccharomyces cerevisiae.
Mellor J, Morillon A. Mellor J, et al. Biochim Biophys Acta. 2004 Mar 15;1677(1-3):100-12. doi: 10.1016/j.bbaexp.2003.10.014. Biochim Biophys Acta. 2004. PMID: 15020051 Review.
Cited by
- Spt5 orchestrates cryptic transcript suppression and transcriptional directionality.
An H, Yang H, Lee D. An H, et al. Commun Biol. 2024 Oct 22;7(1):1370. doi: 10.1038/s42003-024-07014-7. Commun Biol. 2024. PMID: 39438667 Free PMC article. - Dysregulation of epigenetic modifications in inborn errors of immunity.
Xiao Z, He R, Zhao Z, Chen T, Ying Z. Xiao Z, et al. Epigenomics. 2024;16(19-20):1301-1313. doi: 10.1080/17501911.2024.2410695. Epub 2024 Oct 15. Epigenomics. 2024. PMID: 39404224 Review. - Histone variant macroH2A1 regulates synchronous firing of replication origins in the inactive X chromosome.
Arroyo M, Casas-Delucchi CS, Pabba MK, Prorok P, Pradhan SK, Rausch C, Lehmkuhl A, Maiser A, Buschbeck M, Pasque V, Bernstein E, Luck K, Cardoso MC. Arroyo M, et al. Nucleic Acids Res. 2024 Oct 28;52(19):11659-11688. doi: 10.1093/nar/gkae734. Nucleic Acids Res. 2024. PMID: 39189450 Free PMC article. - Swi/Snf chromatin remodeling regulates transcriptional interference and gene repression.
Morse K, Bishop AL, Swerdlow S, Leslie JM, Ünal E. Morse K, et al. Mol Cell. 2024 Aug 22;84(16):3080-3097.e9. doi: 10.1016/j.molcel.2024.06.029. Epub 2024 Jul 22. Mol Cell. 2024. PMID: 39043178 - Temperature-dependent jumonji demethylase modulates flowering time by targeting H3K36me2/3 in Brassica rapa.
Xin X, Li P, Zhao X, Yu Y, Wang W, Jin G, Wang J, Sun L, Zhang D, Zhang F, Yu S, Su T. Xin X, et al. Nat Commun. 2024 Jun 28;15(1):5470. doi: 10.1038/s41467-024-49721-z. Nat Commun. 2024. PMID: 38937441 Free PMC article.
References
- Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–8. - PubMed
- Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. - PubMed
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. - PubMed
- Keogh MC, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases