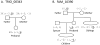A direct characterization of human mutation based on microsatellites - PubMed (original) (raw)
. 2012 Oct;44(10):1161-5.
doi: 10.1038/ng.2398. Epub 2012 Aug 23.
Affiliations
- PMID: 22922873
- PMCID: PMC3459271
- DOI: 10.1038/ng.2398
A direct characterization of human mutation based on microsatellites
James X Sun et al. Nat Genet. 2012 Oct.
Abstract
Mutations are the raw material of evolution but have been difficult to study directly. We report the largest study of new mutations to date, comprising 2,058 germline changes discovered by analyzing 85,289 Icelanders at 2,477 microsatellites. The paternal-to-maternal mutation rate ratio is 3.3, and the rate in fathers doubles from age 20 to 58, whereas there is no association with age in mothers. Longer microsatellite alleles are more mutagenic and tend to decrease in length, whereas the opposite is seen for shorter alleles. We use these empirical observations to build a model that we apply to individuals for whom we have both genome sequence and microsatellite data, allowing us to estimate key parameters of evolution without calibration to the fossil record. We infer that the sequence mutation rate is 1.4-2.3×10(-8) mutations per base pair per generation (90% credible interval) and that human-chimpanzee speciation occurred 3.7-6.6 million years ago.
Figures
Figure 1. Examples of verified mutations from a trio and a family
The proband is the individual inheriting a mutation, and all individuals are named relative to the proband. All alleles are given in repeat units and shifted so that the ancestral allele has length 0. The mutating allele is underlined. (A) We show a mutation detected using the trio approach. Confirmation of the mutation is from multiple genotyping of the trio: the father, mother, and proband are genotyped 3×, 3×, and 4×, respectively. (B) We show a mutation detected using the family approach. One sibling verified the ancestral allele, and one child verified the mutant allele. The phasing of alleles from the mutant locus and other loci from the same chromosome shows that the sibling with alleles (0,-2) did not inherit the ancestral ‘0’ but rather the other ‘0’ allele from the father.
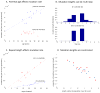
Figure 2. Characteristics of the microsatellite mutation process
(A) Paternal (blue) and maternal (red) mutation rates. The x-axis shows the parental age at child-birth. The data points are grouped into 10 bins (vertical bars show 1 standard error). The paternal rate shows a positive correlation with age (logistic regression of raw data: P=9.3×10−5; slope = 1.1×10−5/yr), with an estimated doubling of rate from age 20 to 58. The maternal rate shows no evidence of increasing with age (P=0.47). (B) Mutation length distributions differ between di- and tetra-nucleotides (upper and lower histograms), with the x-axis in units of step-size. While the di-nucleotide loci experience multi-step mutations in 32% of instances, tetra-nucleotides mutate almost exclusively by a single-step of 4 bases. (C) Mutation rate increases with allele length: di-nucleotides (blue) have a slope of 1.65×10−5per repeat unit (P=1.3×10−3) and tetra-nucleotides (red) have a slope of 6.73×10−5 per repeat unit (P=1.8×10−3). (D) Constraints on allele lengths: When the parental allele is relatively short, mutations tend to increase in length, and when the parental allele is relatively long, mutations tend to decrease in length. Di- and tetra- nucleotides are shown in blue crosses and red circles, respectively. Probit regression of the combined di- and tetra- data shows highly significant evidence of an effect (P=2.8×10−18).
Figure 3. Empirical validation of our model with sequence-based estimates of TMRCA
In red is the simulation of ASD as a function of TMRCA for the standard random walk (GSMM) model. In blue is the simulation of our model, in which the non-linearity compared to GSMM is primarily due to the length constraint that we empirically observed in microsatellites. In black is the empirically observed ASD at microsatellites in 23 HapMap individuals as a function of sequence-based estimates of TMRCA, which is estimated using θseq2μseq, where θseq is the local sequence diversity surrounding each microsatellite locus, and μseq is 1.82×10−8 (obtained from Table 2). The close match of the empirical curve to our model simulations suggests that our model works, and motivates the analysis in which we use the sequence substitution rate in small windows around the microsatellites to make inferences about evolutionary parameters like the sequence mutation rate.
Figure 4. Human-chimpanzee speciation date inferred without a fossil calibration
In the square panel, we give the 90% Bayesian credible interval for human-chimpanzee speciation time (gray), for a range of values of the ratio of speciation time to divergence time τHC/tHC. The blue curve shows our prior probability distribution for τHC/tHC, justified in Supplementary Note. The red horizontal lines are the dates of fossils that are candidates for being on the hominin lineage post-dating the speciation of humans and chimpanzees. Australopithecus amanensis, Orrorin tugenensis and Ardipithecus kadabba are within our plausible speciation times, while Sahelanthropus tchadensis pre-dates the inferred speciation time for all plausible values of τHC/tHC. Our prior distribution for τHC/tHC is shown in the bottom histogram, and our posterior distribution of human-chimpanzee speciation time is shown in the left histogram.
Similar articles
- Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs.
Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A. Kayser M, et al. Am J Hum Genet. 2000 May;66(5):1580-8. doi: 10.1086/302905. Epub 2000 Apr 6. Am J Hum Genet. 2000. PMID: 10762544 Free PMC article. - Microsatellite evolutionary rate and pattern in Schistocerca gregaria inferred from direct observation of germline mutations.
Chapuis MP, Plantamp C, Streiff R, Blondin L, Piou C. Chapuis MP, et al. Mol Ecol. 2015 Dec;24(24):6107-19. doi: 10.1111/mec.13465. Epub 2015 Dec 7. Mol Ecol. 2015. PMID: 26562076 - Markov chain Monte Carlo analysis of human Y-chromosome microsatellites provides evidence of biased mutation.
Cooper G, Burroughs NJ, Rand DA, Rubinsztein DC, Amos W. Cooper G, et al. Proc Natl Acad Sci U S A. 1999 Oct 12;96(21):11916-21. doi: 10.1073/pnas.96.21.11916. Proc Natl Acad Sci U S A. 1999. PMID: 10518551 Free PMC article. - Microsatellite mutations in the germline: implications for evolutionary inference.
Ellegren H. Ellegren H. Trends Genet. 2000 Dec;16(12):551-8. doi: 10.1016/s0168-9525(00)02139-9. Trends Genet. 2000. PMID: 11102705 Review. - Microsatellites: evolution and mutational processes.
Freimer NB, Slatkin M. Freimer NB, et al. Ciba Found Symp. 1996;197:51-67; discussion 67-72. doi: 10.1002/9780470514887.ch4. Ciba Found Symp. 1996. PMID: 8827368 Review.
Cited by
- Characterization of C9orf72 haplotypes to evaluate the effects of normal and pathological variations on its expression and splicing.
Ben-Dor I, Pacut C, Nevo Y, Feldman EL, Reubinoff BE. Ben-Dor I, et al. PLoS Genet. 2021 Mar 29;17(3):e1009445. doi: 10.1371/journal.pgen.1009445. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33780440 Free PMC article. - Short Tandem Repeats in the era of next-generation sequencing: from historical loci to population databases.
Uguen K, Michaud JL, Génin E. Uguen K, et al. Eur J Hum Genet. 2024 Sep;32(9):1037-1044. doi: 10.1038/s41431-024-01666-z. Epub 2024 Jul 10. Eur J Hum Genet. 2024. PMID: 38982300 Review. - Mature microsatellites: mechanisms underlying dinucleotide microsatellite mutational biases in human cells.
Baptiste BA, Ananda G, Strubczewski N, Lutzkanin A, Khoo SJ, Srikanth A, Kim N, Makova KD, Krasilnikova MM, Eckert KA. Baptiste BA, et al. G3 (Bethesda). 2013 Mar;3(3):451-63. doi: 10.1534/g3.112.005173. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450065 Free PMC article. - Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution.
Langergraber KE, Prüfer K, Rowney C, Boesch C, Crockford C, Fawcett K, Inoue E, Inoue-Muruyama M, Mitani JC, Muller MN, Robbins MM, Schubert G, Stoinski TS, Viola B, Watts D, Wittig RM, Wrangham RW, Zuberbühler K, Pääbo S, Vigilant L. Langergraber KE, et al. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15716-21. doi: 10.1073/pnas.1211740109. Epub 2012 Aug 13. Proc Natl Acad Sci U S A. 2012. PMID: 22891323 Free PMC article. Review. - Estimating the human mutation rate using autozygosity in a founder population.
Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, Vives L, O'Roak BJ, Sudmant PH, Shendure J, Abney M, Ober C, Eichler EE. Campbell CD, et al. Nat Genet. 2012 Nov;44(11):1277-81. doi: 10.1038/ng.2418. Epub 2012 Sep 23. Nat Genet. 2012. PMID: 23001126 Free PMC article.
References
- Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–7. - PubMed
- Crow JF. Age and sex effects on human mutation rates: an old problem with new complexities. J Radiat Res (Tokyo) 2006;47 (Suppl B):B75–82. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources