Genome sequencing identifies a basis for everolimus sensitivity - PubMed (original) (raw)
. 2012 Oct 12;338(6104):221.
doi: 10.1126/science.1226344. Epub 2012 Aug 23.
Aphrothiti J Hanrahan, Matthew I Milowsky, Hikmat Al-Ahmadie, Sasinya N Scott, Manickam Janakiraman, Mono Pirun, Chris Sander, Nicholas D Socci, Irina Ostrovnaya, Agnes Viale, Adriana Heguy, Luke Peng, Timothy A Chan, Bernard Bochner, Dean F Bajorin, Michael F Berger, Barry S Taylor, David B Solit
Affiliations
- PMID: 22923433
- PMCID: PMC3633467
- DOI: 10.1126/science.1226344
Genome sequencing identifies a basis for everolimus sensitivity
Gopa Iyer et al. Science. 2012.
Abstract
Cancer drugs often induce dramatic responses in a small minority of patients. We used whole-genome sequencing to investigate the genetic basis of a durable remission of metastatic bladder cancer in a patient treated with everolimus, a drug that inhibits the mTOR (mammalian target of rapamycin) signaling pathway. Among the somatic mutations was a loss-of-function mutation in TSC1 (tuberous sclerosis complex 1), a regulator of mTOR pathway activation. Targeted sequencing revealed TSC1 mutations in about 8% of 109 additional bladder cancers examined, and TSC1 mutation correlated with everolimus sensitivity. These results demonstrate the feasibility of using whole-genome sequencing in the clinical setting to identify previously occult biomarkers of drug sensitivity that can aid in the identification of patients most likely to respond to targeted anticancer drugs.
Trial registration: ClinicalTrials.gov NCT00805129.
Figures
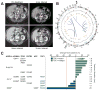
Fig. 1
(A) Computed tomography images of the index patient demonstrating complete resolution of metastatic disease (arrows). (B) Somatic abnormalities in the outlier responder’s genome included (from outside to inside) copy number alterations; mutations at ~10-Mb resolution; regulatory, synonymous, missense, nonsense, nonstop, and frameshift insertion and deletion mutations (black, orange, red, green, and dark green); and intra- and interchromosomal rearrangements (light and dark blue). (C) Best overall response of 14 sequenced trial patients. Negative values indicate tumor shrinkage (red line, threshold for partial response). Gradient arrow, patient with rapid progression in bone.
Comment in
- Genetics: Finding the path to everolimus sensitivity.
Villanueva MT. Villanueva MT. Nat Rev Clin Oncol. 2012 Nov;9(11):609. doi: 10.1038/nrclinonc.2012.163. Epub 2012 Sep 18. Nat Rev Clin Oncol. 2012. PMID: 22987104 No abstract available. - Words of wisdom: Re: Genome sequencing identifies a basis for everolimus sensitivity.
Zlotta AR. Zlotta AR. Eur Urol. 2013 Sep;64(3):516. doi: 10.1016/j.eururo.2013.06.031. Eur Urol. 2013. PMID: 23915465 No abstract available.
Similar articles
- Response and acquired resistance to everolimus in anaplastic thyroid cancer.
Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA, Guo Y, Swanson SJ, Ruan DT, Hanna GJ, Haddad RI, Getz G, Kwiatkowski DJ, Carter SL, Sabatini DM, Jänne PA, Garraway LA, Lorch JH. Wagle N, et al. N Engl J Med. 2014 Oct 9;371(15):1426-33. doi: 10.1056/NEJMoa1403352. N Engl J Med. 2014. PMID: 25295501 Free PMC article. - Everolimus tablets for patients with subependymal giant cell astrocytoma.
Turner SG, Peters KB, Vredenburgh JJ, Desjardins A, Friedman HS, Reardon DA. Turner SG, et al. Expert Opin Pharmacother. 2011 Oct;12(14):2265-9. doi: 10.1517/14656566.2011.601742. Epub 2011 Aug 1. Expert Opin Pharmacother. 2011. PMID: 21806479 Free PMC article. Review. - Next-generation sequencing reveals somatic mutations that confer exceptional response to everolimus.
Lim SM, Park HS, Kim S, Kim S, Ali SM, Greenbowe JR, Yang IS, Kwon NJ, Lee JL, Ryu MH, Ahn JH, Lee J, Lee MG, Kim HS, Kim H, Kim HR, Moon YW, Chung HC, Kim JH, Kang YK, Cho BC. Lim SM, et al. Oncotarget. 2016 Mar 1;7(9):10547-56. doi: 10.18632/oncotarget.7234. Oncotarget. 2016. PMID: 26859683 Free PMC article. - Words of wisdom: Re: Genome sequencing identifies a basis for everolimus sensitivity.
Zlotta AR. Zlotta AR. Eur Urol. 2013 Sep;64(3):516. doi: 10.1016/j.eururo.2013.06.031. Eur Urol. 2013. PMID: 23915465 No abstract available. - Differentiating the mTOR inhibitors everolimus and sirolimus in the treatment of tuberous sclerosis complex.
MacKeigan JP, Krueger DA. MacKeigan JP, et al. Neuro Oncol. 2015 Dec;17(12):1550-9. doi: 10.1093/neuonc/nov152. Epub 2015 Aug 19. Neuro Oncol. 2015. PMID: 26289591 Free PMC article. Review.
Cited by
- Pancreatic cancer: from state-of-the-art treatments to promising novel therapies.
Garrido-Laguna I, Hidalgo M. Garrido-Laguna I, et al. Nat Rev Clin Oncol. 2015 Jun;12(6):319-34. doi: 10.1038/nrclinonc.2015.53. Epub 2015 Mar 31. Nat Rev Clin Oncol. 2015. PMID: 25824606 Review. - Novel molecular targets for urothelial carcinoma.
Faltas BM, Karir BS, Tagawa ST, Rosenberg JE. Faltas BM, et al. Expert Opin Ther Targets. 2015 Apr;19(4):515-25. doi: 10.1517/14728222.2014.987662. Epub 2015 Jan 30. Expert Opin Ther Targets. 2015. PMID: 25633079 Free PMC article. Review. - The phosphoinositide-3-kinase-Akt-mTOR pathway as a therapeutic target in breast cancer.
Lauring J, Park BH, Wolff AC. Lauring J, et al. J Natl Compr Canc Netw. 2013 Jun 1;11(6):670-8. doi: 10.6004/jnccn.2013.0086. J Natl Compr Canc Netw. 2013. PMID: 23744866 Free PMC article. Review. - iGMDR: Integrated Pharmacogenetic Resource Guide to Cancer Therapy and Research.
Chen X, Guo Y, Chen X. Chen X, et al. Genomics Proteomics Bioinformatics. 2020 Apr;18(2):150-160. doi: 10.1016/j.gpb.2019.11.011. Epub 2020 Sep 8. Genomics Proteomics Bioinformatics. 2020. PMID: 32916316 Free PMC article. - Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype.
Phoenix TN, Patmore DM, Boop S, Boulos N, Jacus MO, Patel YT, Roussel MF, Finkelstein D, Goumnerova L, Perreault S, Wadhwa E, Cho YJ, Stewart CF, Gilbertson RJ. Phoenix TN, et al. Cancer Cell. 2016 Apr 11;29(4):508-522. doi: 10.1016/j.ccell.2016.03.002. Epub 2016 Mar 31. Cancer Cell. 2016. PMID: 27050100 Free PMC article.
References
- Materials and methods are available as supplementary materials on Science Online.
- Platt FM, et al. Clin Cancer Res. 2009;15:6008. - PubMed
- Krueger DA, et al. N Engl J Med. 2010;363:1801. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous