Disentangling the many layers of eukaryotic transcriptional regulation - PubMed (original) (raw)
Review
Disentangling the many layers of eukaryotic transcriptional regulation
Katherine M Lelli et al. Annu Rev Genet. 2012.
Abstract
Regulation of gene expression in eukaryotes is an extremely complex process. In this review, we break down several critical steps, emphasizing new data and techniques that have expanded current gene regulatory models. We begin at the level of DNA sequence where cis-regulatory modules (CRMs) provide important regulatory information in the form of transcription factor (TF) binding sites. In this respect, CRMs function as instructional platforms for the assembly of gene regulatory complexes. We discuss multiple mechanisms controlling complex assembly, including cooperative DNA binding, combinatorial codes, and CRM architecture. The second section of this review places CRM assembly in the context of nucleosomes and condensed chromatin. We discuss how DNA accessibility and histone modifications contribute to TF function. Lastly, new advances in chromosomal mapping techniques have provided increased understanding of intra- and interchromosomal interactions. We discuss how these topological maps influence gene regulatory models.
Figures
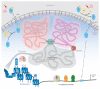
Figure 1
Overview of eukaryotic gene regulation. Within the nucleus of a cell, chromosomes occupy defined spatial regions called territories. Interactions between adjacent territories can correlate with transcriptionally active chromatin in transcription factories or silenced chromatin in Polycomb Group (PcG) bodies. Posttranslational modification of transcription factors (TFs), such as phosphorylation (P), can influence nuclear import (dashed arrows) through nuclear pore complexes (NPCs) in response to extracellular signals. Posttranslational modifications of histones, such as methylation (Me), can also correlate with the transcriptional state of associated genes. The position of nucleosomes can restrict access of TFs by occluding binding sites (colored rectangles). Lastly, TF recognition of specific binding sites, either as monomers or as a part of a complex with other proteins, also contributes to proper recruitment or release of RNA polymerase from the transcriptional start site (TSS). Abbreviations: K, kinase; Ph, phosphatase.
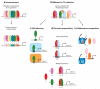
Figure 2
_Cis_-regulatory module (CRM) assembly and cooperative DNA-binding models. This image depicts three different models of CRM assembly and related cooperativity mechanisms. (a) The enhanceosome model requires strict modular cooperativity between all transcription factors (TFs) (129, 131). (b) In contrast, the flexibility of the Billboard and TF Collective models permits different cooperativity mechanisms to control CRM assembly. (c) In the case of DNA allostery, interactions between the DNA sequence and the TF can facilitate conformational changes in the TF that result in the recruitment of different regulatory complexes (rounded rectangles) in a sequence-specific manner (108). (e) Classical cooperativity uses protein-protein interactions between TFs to facilitate cooperative binding. These types of cooperative interactions help to increase TF DNA-binding specificity by restricting recruitment to dimeric sites (A+B). In the case of latent specificity, direct protein-protein interactions alter binding specificities so that TFs recognize novel composite sites (A’B’) (161). (f) Lastly, collaborative competition between TFs and nucleosomes can lead to cooperative binding when the binding of one TF provides access for another TF to bind a neighboring site (114, 120, 167).
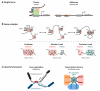
Figure 3
Different types of long-range interactions. (a) Interactions between elements in a single locus. Tissue-specific conformations of elements within the Distalless (Dll) locus correlate with gene activity. In the thorax, where Dll is expressed, the locus is more compact, with several regions interacting over long distances. However, in the abdomen, where Dll is repressed, the locus adapts a more extended conformation, and no interactions are observed between regulatory elements (2). (b) Interactions between elements in a gene complex. In Drosophila three gene complexes, antennapedia (ANT-C), bithorax (BX-C), and NK homeobox (NK-C), create foci of extensive intracomplex interactions when repressed by polycombs called Polycomb group (PcG) bodies (9). Additionally, intercomplex interactions suggest that PcG bodies can encompass more than one complex (9). However, PcG bodies containing all three complexes have not been observed (9). During vertebrate development, Hox genes cluster according to gene activity (125). In the forebrain, where none of the Hox genes are expressed, chromosome conformation capture-on-chip analysis indicates that all members of the complex group together (red). However, in trunk regions Hox genes adopt a bimodal distribution in which all the active genes (green) are found in one cluster and the inactive ones (red) interact within a different cluster (125). Furthermore, this three-dimensional organization correlates with collinear gene activation (125). (c) Interactions between chromosomes. During odorant receptor choice in mouse neurons, trans interactions can occur between the H enhancer and genes on different chromosomes (94). In this case, we have depicted interactions between the H enhancer on chromosome 14 and the M50 gene on chromosome 7 (94). Additionally, interactions between _Klf1_-regulated genes (green) on different chromosomes have been observed in erythroid cells (151). This colocalization of active genes constitutes a transcription factory.
Similar articles
- Advances in analysis of transcriptional regulatory networks.
Kim TM, Park PJ. Kim TM, et al. Wiley Interdiscip Rev Syst Biol Med. 2011 Jan-Feb;3(1):21-35. doi: 10.1002/wsbm.105. Wiley Interdiscip Rev Syst Biol Med. 2011. PMID: 21069662 Review. - The role of chromatin during transcription.
Li B, Carey M, Workman JL. Li B, et al. Cell. 2007 Feb 23;128(4):707-19. doi: 10.1016/j.cell.2007.01.015. Cell. 2007. PMID: 17320508 Review. - De novo prediction of cis-regulatory elements and modules through integrative analysis of a large number of ChIP datasets.
Niu M, Tabari ES, Su Z. Niu M, et al. BMC Genomics. 2014 Dec 2;15:1047. doi: 10.1186/1471-2164-15-1047. BMC Genomics. 2014. PMID: 25442502 Free PMC article. - [Regulatory transcription codes in eukaryotic genomes].
Merkulova TI, Ananko EA, Ignat'eva EV, Kolchanov NA. Merkulova TI, et al. Genetika. 2013 Jan;49(1):37-54. doi: 10.7868/s0016675813010074. Genetika. 2013. PMID: 23662423 Review. Russian. - The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding.
Barrera LO, Ren B. Barrera LO, et al. Curr Opin Cell Biol. 2006 Jun;18(3):291-8. doi: 10.1016/j.ceb.2006.04.002. Epub 2006 May 2. Curr Opin Cell Biol. 2006. PMID: 16647254 Review.
Cited by
- A novel enhancer RNA, Hmrhl, positively regulates its host gene, phkb, in chronic myelogenous leukemia.
Fatima R, Choudhury SR, T R D, Bhaduri U, Rao MRS. Fatima R, et al. Noncoding RNA Res. 2019 Aug 8;4(3):96-108. doi: 10.1016/j.ncrna.2019.08.001. eCollection 2019 Sep. Noncoding RNA Res. 2019. PMID: 31891018 Free PMC article. - Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning.
Slattery M, Voutev R, Ma L, Nègre N, White KP, Mann RS. Slattery M, et al. PLoS Genet. 2013;9(9):e1003753. doi: 10.1371/journal.pgen.1003753. Epub 2013 Sep 5. PLoS Genet. 2013. PMID: 24039600 Free PMC article. - In pursuit of design principles of regulatory sequences.
Levo M, Segal E. Levo M, et al. Nat Rev Genet. 2014 Jul;15(7):453-68. doi: 10.1038/nrg3684. Epub 2014 Jun 10. Nat Rev Genet. 2014. PMID: 24913666 Review. - Evolving insights on how cytosine methylation affects protein-DNA binding.
Dantas Machado AC, Zhou T, Rao S, Goel P, Rastogi C, Lazarovici A, Bussemaker HJ, Rohs R. Dantas Machado AC, et al. Brief Funct Genomics. 2015 Jan;14(1):61-73. doi: 10.1093/bfgp/elu040. Epub 2014 Oct 14. Brief Funct Genomics. 2015. PMID: 25319759 Free PMC article. Review. - Absence of a simple code: how transcription factors read the genome.
Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Slattery M, et al. Trends Biochem Sci. 2014 Sep;39(9):381-99. doi: 10.1016/j.tibs.2014.07.002. Epub 2014 Aug 14. Trends Biochem Sci. 2014. PMID: 25129887 Free PMC article. Review.
References
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell Biochem. 2005;94:890–98. - PubMed
- Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054510/GM/NIGMS NIH HHS/United States
- T32 GM007088/GM/NIGMS NIH HHS/United States
- GM54510/GM/NIGMS NIH HHS/United States
- T32 DK007328/DK/NIDDK NIH HHS/United States
- 5T32DK07328/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous