Transcriptional architecture and chromatin landscape of the core circadian clock in mammals - PubMed (original) (raw)
Transcriptional architecture and chromatin landscape of the core circadian clock in mammals
Nobuya Koike et al. Science. 2012.
Abstract
The mammalian circadian clock involves a transcriptional feed back loop in which CLOCK and BMAL1 activate the Period and Cryptochrome genes, which then feedback and repress their own transcription. We have interrogated the transcriptional architecture of the circadian transcriptional regulatory loop on a genome scale in mouse liver and find a stereotyped, time-dependent pattern of transcription factor binding, RNA polymerase II (RNAPII) recruitment, RNA expression, and chromatin states. We find that the circadian transcriptional cycle of the clock consists of three distinct phases: a poised state, a coordinated de novo transcriptional activation state, and a repressed state. Only 22% of messenger RNA (mRNA) cycling genes are driven by de novo transcription, suggesting that both transcriptional and posttranscriptional mechanisms underlie the mammalian circadian clock. We also find that circadian modulation of RNAPII recruitment and chromatin remodeling occurs on a genome-wide scale far greater than that seen previously by gene expression profiling.
Figures
Fig. 1
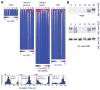
ChIP-seq analysis of the core circadian transcriptional regulators in mouse liver. (A) UCSC genome browser view of BMAL1 (blue), CLOCK (light green), NPAS2 (green), PER1 (orange), PER2 (gold), CRY1 (red) and CRY2 (purple) occupancy at the Dbp (A) locus. Each track represents the normalized ChIP-seq read coverage (wiggle plot) at a single time point. For each transcription factor, six time points every 4 hr over a circadian cycle are shown beginning at CT0 and ending at CT20. Knockout (KO) mice were used as a negative control for each factor except NPAS2. The conservation track shows 30-Way Multiz Alignment & Conservation scores (PhastCons) provided by the UCSC genome browser. Read numbers are normalized as described in the methods. The y-axis scales between tracks for Dbp are BMAL1 (0–70), CLOCK (0–40), NPAS2 (0–25), PER1 (0–35), PER2 (0–60), CRY1 (0–135), CRY2 (0–45). (B) Heat map views of genome-wide DNA binding for BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 measured over 500 bp fragments encompassing the binding sites. Each peak in the genome is represented as a horizontal line, ordered vertically by signal strength. Six time points are shown beginning at CT0 and ending at CT20 from left to right. Knockout (K) mouse control is shown on the far right of each panel. The number of peaks in the genome is indicated at the bottom of each panel. The blue-red gradient indicates the coverage or signal strength (normalized uniquely mapped reads per 10 million reads) for all binding sites in the genome. (C) Histogram showing circadian phase distributions of BMAL1, CLOCK, NPAS2, PER1, PER2, CRY1 and CRY2 binding rhythms. Cyclic binding was determined by ARSER (p<0.05) and the mean circular phases of peak binding as indicated in red are indicated using Oriana. The number of cycling peaks is indicated in black. (D) Chow-Ruskey diagram showing the 6-way overlap of BMAL1, CLOCK, PER1, PER2, CRY1 and CRY2 peaks. Master peak lists were used to determine peak overlaps and an overlap was called if two peak summits were within 120 bp of each other. The red circle in the middle represents the overlap of all six factors. Lighter shades of red, orange and yellow represent fewer overlaps of subsets. The boundaries for each protein are color coded: BMAL1 (blue), CLOCK (green), PER1 (orange), PER2 (brown), CRY1 (red) and CRY2 (purple), and the areas of each domain are proportional to the number of binding sites.
Fig. 2
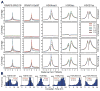
Whole-transcriptome RNA-seq analysis of circadian gene expression. (A) UCSC genome browser views of RNA-seq data at the Per2 locus. Sense strand reads are shown in black and reverse strand reads in red as normalized average reads per 50 million total reads in 10 bp bins. The y-axis scales are: Per2 (0–40 for the positive strand, −40–0 for the negative strand). (B) RNA-seq read coverage in reads per kilobase per million (RPKM) reads in exons and introns. The intron (blue) and exon (red) RNA expression of Per2 is circadian (ARSER: p<0.01). (C) Cyclic expression of Per2 sense and antisense transcripts. The Per2 antisense transcript is circadian (ARSER; p<0.05) and antiphasic (phase of sense: CT14.4; phase of antisense: CT7.1). (D) Heat map view of intron (left) and exon (right) cycling genes. Each gene is represented as a horizontal line, ordered vertically by phase determined by ARSER. (E) The phase distribution of cycling genes. The phase of each transcript rhythm is represented in a dot plot (top), rose plot (middle) and histogram plot (bottom). The mean circular phase of the rhythms is indicated in red. (F) Venn diagram of intron and exon cycling genes. (G) Comparison of the phase of intron and exon cycling transcripts for common genes. The RNA peak phase of intron (blue) and exon (red) is double plotted and ordered by intron phase. (H) Histogram of intron phase in intron-only cycling genes, intron phase in common genes, exon phase in common genes, and exon phase in exon-only cycling genes (mean circular phase is shown in red).
Fig. 3
Circadian expression of RNAPII and co-activator occupancy. (A) Heat map view of genome-wide DNA binding for p300, RNAPII-8WG16, RNAPII-Ser5P and CBP as described in Fig. 1B. Histograms of the phase of the rhythms are shown below the heat map (mean circular phase is shown in red and number of cycling peaks is indicated in black). (B) PER2 and CBP protein interaction using co-immunoprecipitation of native extracts during the circadian cycle. Co-immunoprecipitation was carried out with anti-CBP antibody and mouse cerebellum protein lysate. The immunoprecipitates were visualized on western blots with anti-PER2 or anti-CBP antibodies.
Fig. 4
Circadian regulation of histone modifications. (A) Binding profile of RNAPII-8WG16, RNAPII-Ser5P, H3K4me3, H3K9ac and H3K27ac at the TSS +/− 3kb in 12,680 expressed genes (top row), 8,945 unexpressed genes (second row), 1371 intron cycling genes (third row) and 5,839 non-cycling genes (bottom row). The y-axis represents the average signal in a 10 bp bin (normalized uniquely mapped reads per 10 million reads). (B) Histograms showing circadian phase distributions of histone modification rhythms (ARSER: p<0.05). Mean circular phase is shown in red. The signal in TSS +/− 1 kb was used to determine H3K4me1, H3K4me3, H3K9ac and H3K27ac binding rhythm phases. The signal in the gene body was used for H3K36me3 and H3K79me2. Genes without MACS peaks in the corresponding area (TSS or gene body) were filtered out from the analysis.
Fig. 5
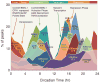
Circadian landscape of the cistrome and epigenome of the liver. Phase distributions of circadian transcriptional regulators, intron cycling RNA transcripts and histone modifications as shown in Fig. 1C, 2E, 3A and 4B. The mean circular phase of peak binding is indicated under the name.
Comment in
- Circadian surprise--it's not all about transcription.
Doherty CJ, Kay SA. Doherty CJ, et al. Science. 2012 Oct 19;338(6105):338-40. doi: 10.1126/science.1230008. Science. 2012. PMID: 23087238 No abstract available.
Similar articles
- Molecular components of the circadian clock in mammals.
Takahashi JS. Takahashi JS. Diabetes Obes Metab. 2015 Sep;17 Suppl 1(0 1):6-11. doi: 10.1111/dom.12514. Diabetes Obes Metab. 2015. PMID: 26332962 Free PMC article. Review. - Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock.
Ye R, Selby CP, Chiou YY, Ozkan-Dagliyan I, Gaddameedhi S, Sancar A. Ye R, et al. Genes Dev. 2014 Sep 15;28(18):1989-98. doi: 10.1101/gad.249417.114. Genes Dev. 2014. PMID: 25228643 Free PMC article. - Rhythmic histone acetylation underlies transcription in the mammalian circadian clock.
Etchegaray JP, Lee C, Wade PA, Reppert SM. Etchegaray JP, et al. Nature. 2003 Jan 9;421(6919):177-82. doi: 10.1038/nature01314. Epub 2002 Dec 11. Nature. 2003. PMID: 12483227 - Transcriptional architecture of the mammalian circadian clock.
Takahashi JS. Takahashi JS. Nat Rev Genet. 2017 Mar;18(3):164-179. doi: 10.1038/nrg.2016.150. Epub 2016 Dec 19. Nat Rev Genet. 2017. PMID: 27990019 Free PMC article. Review. - Circadian surprise--it's not all about transcription.
Doherty CJ, Kay SA. Doherty CJ, et al. Science. 2012 Oct 19;338(6105):338-40. doi: 10.1126/science.1230008. Science. 2012. PMID: 23087238 No abstract available.
Cited by
- Epigenetic mechanisms in diurnal cycles of metabolism and neurodevelopment.
Powell WT, LaSalle JM. Powell WT, et al. Hum Mol Genet. 2015 Oct 15;24(R1):R1-9. doi: 10.1093/hmg/ddv234. Epub 2015 Jun 23. Hum Mol Genet. 2015. PMID: 26105183 Free PMC article. Review. - Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging.
Wang RH, Zhao T, Cui K, Hu G, Chen Q, Chen W, Wang XW, Soto-Gutierrez A, Zhao K, Deng CX. Wang RH, et al. Sci Rep. 2016 Jun 27;6:28633. doi: 10.1038/srep28633. Sci Rep. 2016. PMID: 27346580 Free PMC article. - Genome-wide circadian regulation: A unique system for computational biology.
Sun L, Ma J, Turck CW, Xu P, Wang GZ. Sun L, et al. Comput Struct Biotechnol J. 2020 Jul 10;18:1914-1924. doi: 10.1016/j.csbj.2020.07.002. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32774786 Free PMC article. Review. - Identifying Transcription Factor Combinations to Modulate Circadian Rhythms by Leveraging Virtual Knockouts on Transcription Networks.
Chowdhury D, Wang C, Lu A, Zhu H. Chowdhury D, et al. iScience. 2020 Aug 22;23(9):101490. doi: 10.1016/j.isci.2020.101490. eCollection 2020 Sep 25. iScience. 2020. PMID: 32920484 Free PMC article. - The Catalytic and Non-catalytic Functions of the Brahma Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in Drosophila.
Kwok RS, Li YH, Lei AJ, Edery I, Chiu JC. Kwok RS, et al. PLoS Genet. 2015 Jul 1;11(7):e1005307. doi: 10.1371/journal.pgen.1005307. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26132408 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases