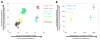A high-coverage genome sequence from an archaic Denisovan individual - PubMed (original) (raw)
. 2012 Oct 12;338(6104):222-6.
doi: 10.1126/science.1224344. Epub 2012 Aug 30.
Martin Kircher, Marie-Theres Gansauge, Heng Li, Fernando Racimo, Swapan Mallick, Joshua G Schraiber, Flora Jay, Kay Prüfer, Cesare de Filippo, Peter H Sudmant, Can Alkan, Qiaomei Fu, Ron Do, Nadin Rohland, Arti Tandon, Michael Siebauer, Richard E Green, Katarzyna Bryc, Adrian W Briggs, Udo Stenzel, Jesse Dabney, Jay Shendure, Jacob Kitzman, Michael F Hammer, Michael V Shunkov, Anatoli P Derevianko, Nick Patterson, Aida M Andrés, Evan E Eichler, Montgomery Slatkin, David Reich, Janet Kelso, Svante Pääbo
Affiliations
- PMID: 22936568
- PMCID: PMC3617501
- DOI: 10.1126/science.1224344
A high-coverage genome sequence from an archaic Denisovan individual
Matthias Meyer et al. Science. 2012.
Abstract
We present a DNA library preparation method that has allowed us to reconstruct a high-coverage (30×) genome sequence of a Denisovan, an extinct relative of Neandertals. The quality of this genome allows a direct estimation of Denisovan heterozygosity indicating that genetic diversity in these archaic hominins was extremely low. It also allows tentative dating of the specimen on the basis of "missing evolution" in its genome, detailed measurements of Denisovan and Neandertal admixture into present-day human populations, and the generation of a near-complete catalog of genetic changes that swept to high frequency in modern humans since their divergence from Denisovans.
Figures
Figure 1
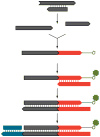
For single-stranded library preparation, ancient DNA molecules are dephosphorylated and heat-denatured. A biotinylated adaptor oligonucleotide is ligated to 3′-ends of the molecules, which are immobilized on streptavidin-coated beads and copied by extension of a primer hybridized to the adaptor. One strand of a double-stranded adaptor is then ligated to the newly synthesized strand. Finally, the beads are destroyed by heat to release the library molecules (not shown).
Figure 2
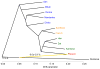
Average sequence divergence and branch length differences between the Denisovan genome and 11 present-day humans represented as a tree. Divergence is reported as fraction of the branch leading from human to the common ancestor with chimpanzee, and in years, assuming a human-chimpanzee divergence of 6.5 million years ago.
Figure 3
Maximum likelihood tree relating the Denisovan genome and the genomes of eleven present-day humans, allowing one migration event (shown in yellow).
Figure 4
(A) Sharing of derived alleles among present-day humans, Denisovans and Neandertals. We compare all possible pairs of 11 present-day humans {H1, H2} in their “_D-_statistics”, which measure the rate at which they share derived alleles with Denisovans (x-axis) and Neandertals (y-axis). Each point reports ±1 standard error bars from a Block Jackknife. _D_-statistics are color-coded by geographic region. The _D-_statistic is not the same as the mixture proportion; it is also affected by quantities like the amount of shared genetic drift between the samples being compared. (B) Sharing of derived alleles that are absent in Africans among present-day humans, Denisovans and Neandertals. We enhance the power of the _D-_statistics by restricting to sites where 35 sub-Saharan African samples have the ancestral allele, and pooling modern humans by region to increase resolution (bars again give one standard error). Eastern non-African populations have significantly more archaic ancestry than European populations (Z=5.3 and Z=4.8 for the tests based on the Denisovan and Neandertal _D-_statistics, respectively).
Figure 5
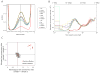
(A) Heterozygosity shown by the distribution of the number of bases matching the human reference genome at sites sampled to 20-fold coverage. The Y-axis is scaled to show the peak representing heterozygous sites in the center. (B) Inference of population size change over time using variation in the time since the most recent common ancestors across the genome shows that Denisovans have had a small population size over the last few hundred thousand years compared with modern humans, but a similar demographic history earlier. The y-axis specifies a number proportional to the population size Ne. The x-axis specifies time in units of divergence per base pair (along the top in years, assuming rates of 0.5×10−9 to 1.0×10−9 per year). Thin red lines around the Denisovan curve represent 100 bootstraps, thus showing the uncertainty of the inference. (C) The small population size in Denisovans is reflected in a greater accumulation of non-synonymous sites (normalized by the number of synonymous sites), whether measured in terms of heterozygous sites in Denisovans vs. modern humans (ratio 2.0 –2.5), or the accumulation of divergent sites on the Denisovan lineage divided by modern human lineages (ratio 1.5–2.0). The analysis is restricted to non-synonymous sites predicted to have a possibly or probably damaging effect on protein structure or function.
Similar articles
- Early history of Neanderthals and Denisovans.
Rogers AR, Bohlender RJ, Huff CD. Rogers AR, et al. Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):9859-9863. doi: 10.1073/pnas.1706426114. Epub 2017 Aug 7. Proc Natl Acad Sci U S A. 2017. PMID: 28784789 Free PMC article. - Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals.
Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, Scheinfeldt LB, Merriwether DA, Koki G, Friedlaender JS, Wakefield J, Pääbo S, Akey JM. Vernot B, et al. Science. 2016 Apr 8;352(6282):235-9. doi: 10.1126/science.aad9416. Epub 2016 Mar 17. Science. 2016. PMID: 26989198 Free PMC article. - Chromosomal Rearrangements as Barriers to Genetic Homogenization between Archaic and Modern Humans.
Rogers RL. Rogers RL. Mol Biol Evol. 2015 Dec;32(12):3064-78. doi: 10.1093/molbev/msv204. Epub 2015 Sep 23. Mol Biol Evol. 2015. PMID: 26399483 Free PMC article. - Archaic hominin admixture and its consequences for modern humans.
Tagore D, Akey JM. Tagore D, et al. Curr Opin Genet Dev. 2025 Feb;90:102280. doi: 10.1016/j.gde.2024.102280. Epub 2024 Nov 21. Curr Opin Genet Dev. 2025. PMID: 39577372 Review. - Archaic human genomics.
Disotell TR. Disotell TR. Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review.
Cited by
- Detecting adaptive introgression in human evolution using convolutional neural networks.
Gower G, Picazo PI, Fumagalli M, Racimo F. Gower G, et al. Elife. 2021 May 25;10:e64669. doi: 10.7554/eLife.64669. Elife. 2021. PMID: 34032215 Free PMC article. - An early modern human from Romania with a recent Neanderthal ancestor.
Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, Viola B, Prüfer K, Meyer M, Kelso J, Reich D, Pääbo S. Fu Q, et al. Nature. 2015 Aug 13;524(7564):216-9. doi: 10.1038/nature14558. Epub 2015 Jun 22. Nature. 2015. PMID: 26098372 Free PMC article. - Mapping gene flow between ancient hominins through demography-aware inference of the ancestral recombination graph.
Hubisz MJ, Williams AL, Siepel A. Hubisz MJ, et al. PLoS Genet. 2020 Aug 6;16(8):e1008895. doi: 10.1371/journal.pgen.1008895. eCollection 2020 Aug. PLoS Genet. 2020. PMID: 32760067 Free PMC article. - Ancient DNA damage.
Dabney J, Meyer M, Pääbo S. Dabney J, et al. Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a012567. doi: 10.1101/cshperspect.a012567. Cold Spring Harb Perspect Biol. 2013. PMID: 23729639 Free PMC article. Review. - The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes).
McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, Fontenot MB, Bradley BJ. McIntosh AM, et al. PLoS One. 2012;7(10):e47760. doi: 10.1371/journal.pone.0047760. Epub 2012 Oct 24. PLoS One. 2012. PMID: 23112842 Free PMC article.
References
- Gibbons A. Paleoanthropology. Who were the Denisovans? Science. 2011;333:1084. - PubMed
Publication types
MeSH terms
Grants and funding
- R01-GM40282/GM/NIGMS NIH HHS/United States
- R01 GM100233/GM/NIGMS NIH HHS/United States
- R01 GM040282/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM100233/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources