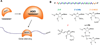Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression - PubMed (original) (raw)
Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression
Dongbo Yu et al. Cell. 2012.
Abstract
Mutant huntingtin (HTT) protein causes Huntington disease (HD), an incurable neurological disorder. Silencing mutant HTT using nucleic acids would eliminate the root cause of HD. Developing nucleic acid drugs is challenging, and an ideal clinical approach to gene silencing would combine the simplicity of single-stranded antisense oligonucleotides with the efficiency of RNAi. Here, we describe RNAi by single-stranded siRNAs (ss-siRNAs). ss-siRNAs are potent (>100-fold more than unmodified RNA) and allele-selective (>30-fold) inhibitors of mutant HTT expression in cells derived from HD patients. Strategic placement of mismatched bases mimics micro-RNA recognition and optimizes discrimination between mutant and wild-type alleles. ss-siRNAs require Argonaute protein and function through the RNAi pathway. Intraventricular infusion of ss-siRNA produced selective silencing of the mutant HTT allele throughout the brain in a mouse HD model. These data demonstrate that chemically modified ss-siRNAs function through the RNAi pathway and provide allele-selective compounds for clinical development.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. AGO-modulated gene silencing and chemically modified ss-siRNAs
(A) Recognition of mRNA by ss-siRNA. ss-siRNA loads into AGO protein and the complex recognizes a target sequence within an mRNA to silence gene expression. (B) Sequence and chemical modifications of a typical ss-siRNA. The 5'-thymidine base is modified with an _(E)_-vinylphosphonate.
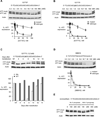
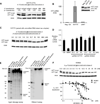
Figure 2. ss-siRNAs inhibit HTT expression
Western analysis of inhibition of HTT expression by: (A) ss-siRNA 537787 (no mismatches); (B) ss-siRNA 537775 (one mismatch at P9). (C) ss-siRNA 537775 over fourteen days with quantitation. (D) a methoxyethyl antisense oligonucleotide that targets a non-repeat region of HTT mRNA. The graphs in parts A,B, and D show amounts of both wild-type and mutant HTT protein. (E) a fully complementary single-stranded RNA lacking any chemical modifications with and without a 5’ terminal phosphate. MM: an RNA duplex containing multiple mismatches. Western analysis is representative data of at least duplicate experiments. Error bars are standard error of the mean (SEM) for dose response studies from three or more independent experiments.
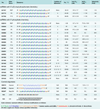
Figure 3. ss-siRNAs and other oligonucleotides used in these studies
N.I.: no inhibition; N/A: Not available. Except for CM (a dsRNA species), all Tm's are measured using ss-siRNAs duplexed with equimolar amounts of unmodified ssRNA 5'- CAGCAGCAGCAGCAGCAGCAGC-3'. Gapmer oligonucleotides have backbones containing only phosphorothioate linkages. All data were obtained using HD patient-derived fibroblast cell line GM04281. Selectivity is calculated by dividing the IC50 of for inhibition of wild-type HTT expression by that for mutant HTT. Error ranges represent standard error of the mean (SEM) of IC50 values from biological replicates.
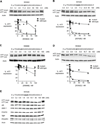
Figure 4. Characterization of inhibition by modified ss-siRNAs
Western analysis of inhibition of HTT expression by: (A) ss-siRNA 553822 (mismatched base at P9) containing a 5'-phosphate; (B) ss-siRNA 557426 containing three central mismatches; (C) ss-siRNA 556888 containing a mismatch at P6; and (D) ss-siRNA 553822 (mismatched base at P9) in 44-CAG-repeat GM04719 cells. (E) Effect of ss-siRNA 537775 on other genes containing trinucleotide repeats. MM: an RNA containing multiple mismatches. Western analysis for parts A–C is representative data from three or more experiments and error bars are standard error of the mean (SEM).
Figure 5. Mechanism of allele-selective inhibition of HTT by ss-siRNA
(A) Western analysis of the effect siRNA-mediated reduction of AGO1-4 expression on allele-selective inhibition by ss-siRNA 537775. (B) RNA immunoprecipitation (RIP) using anti-AGO2 antibody after transfection of ss-siRNA 537775, a control ss-siRNA 522247 not targeting HTT, or an allele-selective single-stranded ASO (LNAT) (Hu et al., 2009) at 25nM. Y-axis measures fold-enrichment of HTT mRNA of anti-AGO2 vs. IgG pulldown. (C) Western analysis of inhibition of HTT expression by ss-siRNA 537775 in complex with a complementary unmodified RNA. (D) Effect of ss-siRNAs 537775 or 553822 on levels of HTT mRNA evaluated by QPCR. (E) In vitro assays using recombinant RNase H and Ago2 proteins do not show efficient substrate cleavage by ss-siRNA. (F) Primary data and Hill plot used for determining cooperativity of HTT inhibition by ss-siRNA 553822. X-axis shows ss-siRNA concentration in logarithmic scale. Hill’s coefficient (nh) is 2.2 ± 0.3 for mutant HTT and 1.2 ± 0.2 for wild-type HTT. Error bars from western quantitation and RIP are standard error of the mean (SEM) from three or more independent experiments, and error bars on HTT mRNA levels are standard deviations (SD) from replicate data. See also Figure S2.
Figure 6. Allele-selective inhibition of HTT by ssiRNA in Q150 HD mouse model
(A) Western analysis of HTT expression on allele-selective inhibition by ss-siRNA 537775 (n=5) in Q150/Q7 mouse frontal cortex. (B) Quantitation of wild-type and mutant HTT protein levels shown in (A). (C) Q-PCR analysis of HTT mRNA levels in mouse frontal cortex after treatment with ss-siRNA, vehicle, or control gapmer ASO. (D) Western analysis HTT expression after allele-selective inhibition by ss-siRNA 537773 (n=5) in different brain regions. (E) Quantitation of western analysis from (D). Results from each treatment group/brain section were averaged. Error bars represent standard error of the mean (SEM) after averaging quantitation results from multiple gel images. *p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S3.
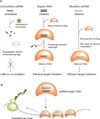
Figure 7. Action of chemically modified ss-siRNAs and allele-selective inhibition of HTT
(A) Chemical modifications allow ss-siRNA to be stable and function through RNAi pathway inside cells. (B) Binding of multiple anti-CAG ss-siRNA:AGO2 complexes to expanded trinucleotide repeat contributes to allele-selective inhibition.
Comment in
- Singles engage the RNA interference pathway.
Davidson BL, Monteys AM. Davidson BL, et al. Cell. 2012 Aug 31;150(5):873-5. doi: 10.1016/j.cell.2012.08.008. Cell. 2012. PMID: 22939614
Similar articles
- Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice.
Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Boudreau RL, et al. Mol Ther. 2009 Jun;17(6):1053-63. doi: 10.1038/mt.2009.17. Epub 2009 Feb 24. Mol Ther. 2009. PMID: 19240687 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo.
Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, Thompson LM, Muchowski PJ. Legleiter J, et al. J Biol Chem. 2010 May 7;285(19):14777-90. doi: 10.1074/jbc.M109.093708. Epub 2010 Mar 10. J Biol Chem. 2010. PMID: 20220138 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. PMID: 28752910 Free PMC article. Updated. Review.
Cited by
- Emerging Oligonucleotide Therapeutics for Rare Neuromuscular Diseases.
Aoki Y, Wood MJA. Aoki Y, et al. J Neuromuscul Dis. 2021;8(6):869-884. doi: 10.3233/JND-200560. J Neuromuscul Dis. 2021. PMID: 34092651 Free PMC article. Review. - Selective enhancing effect of early mitotic inhibitor 1 (Emi1) depletion on the sensitivity of doxorubicin or X-ray treatment in human cancer cells.
Shimizu N, Nakajima NI, Tsunematsu T, Ogawa I, Kawai H, Hirayama R, Fujimori A, Yamada A, Okayasu R, Ishimaru N, Takata T, Kudo Y. Shimizu N, et al. J Biol Chem. 2013 Jun 14;288(24):17238-52. doi: 10.1074/jbc.M112.446351. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645673 Free PMC article. - Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities.
Tabrizi SJ, Flower MD, Ross CA, Wild EJ. Tabrizi SJ, et al. Nat Rev Neurol. 2020 Oct;16(10):529-546. doi: 10.1038/s41582-020-0389-4. Epub 2020 Aug 14. Nat Rev Neurol. 2020. PMID: 32796930 Review. - Limits of using oligonucleotides for allele-selective inhibition at trinucleotide repeat sequences - targeting the CAG repeat within ataxin-1.
Hu J, Corey DR. Hu J, et al. Nucleosides Nucleotides Nucleic Acids. 2020;39(1-3):185-194. doi: 10.1080/15257770.2019.1671592. Epub 2019 Oct 24. Nucleosides Nucleotides Nucleic Acids. 2020. PMID: 31645175 Free PMC article. - RNAi modulation of placental sFLT1 for the treatment of preeclampsia.
Turanov AA, Lo A, Hassler MR, Makris A, Ashar-Patel A, Alterman JF, Coles AH, Haraszti RA, Roux L, Godinho BMDC, Echeverria D, Pears S, Iliopoulos J, Shanmugalingam R, Ogle R, Zsengeller ZK, Hennessy A, Karumanchi SA, Moore MJ, Khvorova A. Turanov AA, et al. Nat Biotechnol. 2018 Nov 19:10.1038/nbt.4297. doi: 10.1038/nbt.4297. Online ahead of print. Nat Biotechnol. 2018. PMID: 30451990 Free PMC article.
References
- Braasch DA, Jensen S, Liu Y, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically modified RNA. Biochemistry. 2003;42:7967–7975. - PubMed
- Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials