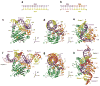Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p - PubMed (original) (raw)
Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p
Anna L Mallam et al. Nature. 2012.
Abstract
DEAD-box proteins are the largest family of nucleic acid helicases, and are crucial to RNA metabolism throughout all domains of life. They contain a conserved 'helicase core' of two RecA-like domains (domains (D)1 and D2), which uses ATP to catalyse the unwinding of short RNA duplexes by non-processive, local strand separation. This mode of action differs from that of translocating helicases and allows DEAD-box proteins to remodel large RNAs and RNA-protein complexes without globally disrupting RNA structure. However, the structural basis for this distinctive mode of RNA unwinding remains unclear. Here, structural, biochemical and genetic analyses of the yeast DEAD-box protein Mss116p indicate that the helicase core domains have modular functions that enable a novel mechanism for RNA-duplex recognition and unwinding. By investigating D1 and D2 individually and together, we find that D1 acts as an ATP-binding domain and D2 functions as an RNA-duplex recognition domain. D2 contains a nucleic-acid-binding pocket that is formed by conserved DEAD-box protein sequence motifs and accommodates A-form but not B-form duplexes, providing a basis for RNA substrate specificity. Upon a conformational change in which the two core domains join to form a 'closed state' with an ATPase active site, conserved motifs in D1 promote the unwinding of duplex substrates bound to D2 by excluding one RNA strand and bending the other. Our results provide a comprehensive structural model for how DEAD-box proteins recognize and unwind RNA duplexes. This model explains key features of DEAD-box protein function and affords a new perspective on how the evolutionarily related cores of other RNA and DNA helicases diverged to use different mechanisms.
Figures
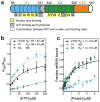
Figure 1. The distinct substrate binding characteristics of the helicase core domains of Mss116p
a, Schematic of the domain architecture of the helicase core of Mss116p (D1, blue; D2, green; C-terminal extension of D2 (CTE), orange), indicating conserved DEAD-box sequence motifs defined according to Fairman-Williams et al. (2010). Full-length Mss116p contains additional unstructured N-terminal (residues 37–87) and C-terminal (residues 598–664) regions that are not required for helicase activity,. b, Affinity of ATP for D1, D2, and the full helicase core (D1D2) measured by gel filtration chromatography under equilibrium conditions. ATP binding was also assessed by an ATP-agarose binding assay (Supplementary Fig. 1). c, Affinity of FAM-dsRNA (Fig. 2a) for MBP-tagged D1, D2 and D1D2 determined by fluorescence anisotropy measurements. Similar results for dsRNA binding were obtained by EMSA (Supplementary Fig. 2). Error bars in b and c represent the standard error for at least three independent measurements, and the error in the _K_d represents the standard error of the non-linear regression (see Full Methods). NB indicates no significant binding.
Figure 2. Crystal structures of Mss116p D2 bound to A-form duplexes
a, The 14-bp self-complementary GC-rich RNA duplex substrate. b, The 14-bp GC-rich chimeric RNA-DNA duplex substrate. c-e, Orthogonal views of the D2-dsRNA complex colored as in Fig. 1a and Fig. 2a. Helix α 14 of D2, which contains motif IVa, faces the major groove of the dsRNA, and α 18 and α 20 of the CTE face the minor groove of the dsRNA. f-h, Orthogonal views of the D2-dsRNA-DNA complex, colored as in Fig. 1a and Fig. 2b, in which D2 is bound to two stacked 14-bp chimeric RNA-DNA duplexes.
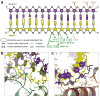
Figure 3. Interactions between Mss116p D2 and duplex RNA
a, Schematic of RNA-protein interactions observed in the D2-dsRNA structure. The dsRNA interacts with conserved DEAD-box motifs IV-Va of D2 (green) and the CTE of D2 (orange). The box indicates that the interaction is maintained in closed-state Mss116p. RNA bases are numbered according to their position relative to ssRNA in closed-state Mss116p (see Supplementary Fig. 4d). Similar nucleic acid-protein interactions were observed in the D2-dsRNA-DNA structure (Supplementary Fig. 5). b, Interactions between strand 1 (yellow) of the duplex RNA and D2 (green). c, Interactions between duplex RNA and the CTE of D2 (orange).
Figure 4. RNA duplex binding and unwinding by Mss116p
a, Model for the modular roles of the helicase core domains of Mss116p during RNA recognition and unwinding. Although ATP and duplex RNA could bind in either order, the binding of ATP to D1 is shown as a first step because Mss116p-ATP complexes are likely pre-populated at physiological concentrations of ATP (>1 mM). b. Comparison of the position and interactions of the flexible motif Va loop (residues 435–440) in the D2-dsRNA and closed-state structures of Mss116p. The closed-state helicase core of Mss116p (PDB = 3I5X) bound to ssRNA (U10-RNA; yellow) and adenosine nucleotide (AMP-PNP; black) is shown with domains colored as in Fig. 1a. In the D2-dsRNA structure, the motif Va loop (green) interacts with strand 1 of dsRNA (yellow), whereas in the closed-state structure, the loop (red) shifts to a different position where motif Va helps to form the ATP-binding site. c, Surface representation of closed-state Mss116p with N1–N10 of U10-RNA (yellow) indicated. d, Surface representation of closed-state Mss116p with dsRNA modeled in the duplex RNA-binding pocket of D2. Sterically incompatible regions of D1 are highlighted in red. e, Change in trajectory of strand 1 and predicted displacement of strand 2 of bound dsRNA by D1 upon core closure of Mss116p with arrows indicating regions of the substrate that are displaced.
Similar articles
- DEAD-box proteins unwind duplexes by local strand separation.
Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. Yang Q, et al. Mol Cell. 2007 Oct 26;28(2):253-63. doi: 10.1016/j.molcel.2007.08.016. Mol Cell. 2007. PMID: 17964264 - Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro.
Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Mohr G, et al. J Mol Biol. 2008 Feb 1;375(5):1344-64. doi: 10.1016/j.jmb.2007.11.041. Epub 2007 Nov 22. J Mol Biol. 2008. PMID: 18096186 Free PMC article. - Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Mallam AL, Sidote DJ, Lambowitz AM. Mallam AL, et al. Elife. 2014 Dec 12;3:e04630. doi: 10.7554/eLife.04630. Elife. 2014. PMID: 25497230 Free PMC article. - The mechanism of ATP-dependent RNA unwinding by DEAD box proteins.
Hilbert M, Karow AR, Klostermeier D. Hilbert M, et al. Biol Chem. 2009 Dec;390(12):1237-50. doi: 10.1515/BC.2009.135. Biol Chem. 2009. PMID: 19747077 Review. - Fluorescence methods in the investigation of the DEAD-box helicase mechanism.
Andreou AZ, Klostermeier D. Andreou AZ, et al. Exp Suppl. 2014;105:161-92. doi: 10.1007/978-3-0348-0856-9_8. Exp Suppl. 2014. PMID: 25095995 Review.
Cited by
- Yeast DEAD box protein Mss116p is a transcription elongation factor that modulates the activity of mitochondrial RNA polymerase.
Markov DA, Wojtas ID, Tessitore K, Henderson S, McAllister WT. Markov DA, et al. Mol Cell Biol. 2014 Jul;34(13):2360-9. doi: 10.1128/MCB.00160-14. Epub 2014 Apr 14. Mol Cell Biol. 2014. PMID: 24732805 Free PMC article. - A long noncoding RNA protects the heart from pathological hypertrophy.
Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP. Han P, et al. Nature. 2014 Oct 2;514(7520):102-106. doi: 10.1038/nature13596. Epub 2014 Aug 10. Nature. 2014. PMID: 25119045 Free PMC article. - RNA folding and functions of RNA helicases in ribosome biogenesis.
Mitterer V, Pertschy B. Mitterer V, et al. RNA Biol. 2022 Jan;19(1):781-810. doi: 10.1080/15476286.2022.2079890. RNA Biol. 2022. PMID: 35678541 Free PMC article. Review. - Concurrent remodelling of nucleolar 60S subunit precursors by the Rea1 ATPase and Spb4 RNA helicase.
Mitterer V, Thoms M, Buschauer R, Berninghausen O, Hurt E, Beckmann R. Mitterer V, et al. Elife. 2023 Mar 17;12:e84877. doi: 10.7554/eLife.84877. Elife. 2023. PMID: 36929751 Free PMC article. - Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate.
Zhou D, Zhu X, Zheng S, Tan D, Dong MQ, Ye K. Zhou D, et al. Protein Cell. 2019 Feb;10(2):120-130. doi: 10.1007/s13238-018-0526-7. Epub 2018 Mar 19. Protein Cell. 2019. PMID: 29557065 Free PMC article.
References
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. - PubMed
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM037951/GM/NIGMS NIH HHS/United States
- R37 GM037951/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM037951/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases